6N6L
 
 | | Crystal Structure of ATPase delta 1-79 Spa47 R189A R191A mutant | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N6Z
 
 | |
6N6M
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R189A | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N70
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R191A | | Descriptor: | ATP synthase SpaL/MxiB | | Authors: | Morales, Y, Olsen, K.J, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N74
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R271E | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N71
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R191E | | Descriptor: | Probable ATP synthase SpaL/MxiB | | Authors: | Morales, Y, Olsen, K.J, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N72
 
 | |
6N73
 
 | |
1QG7
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | STROMAL CELL-DERIVED FACTOR 1 ALPHA, SULFATE ION | | Authors: | Senda, T, Nandhagopal, N, Sugimoto, K, Mitsui, Y. | | Deposit date: | 1999-04-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of recombinant native SDF-1alpha with additional mutagenesis studies: an attempt at a more comprehensive interpretation of accumulated structure-activity relationship data.
J.Interferon Cytokine Res., 20, 2000
|
|
2KT8
 
 | | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B | | Descriptor: | Probable surface protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B
To be Published
|
|
6R2E
 
 | | Crystal structure of the human thymidylate synthase (hTS) interface variant Q62R | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evidence of Destabilization of the Human Thymidylate Synthase (hTS) Dimeric Structure Induced by the Interface Mutation Q62R.
Biomolecules, 9, 2019
|
|
3GRD
 
 | |
2ADI
 
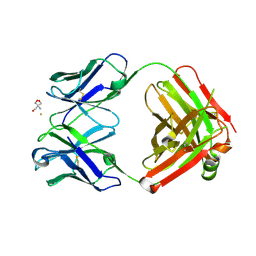 | | Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Barium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BARIUM ION, Q425 Fab Heavy chain, ... | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2ADG
 
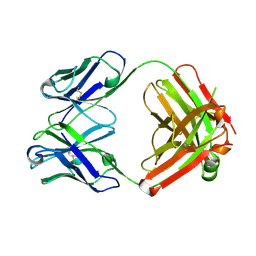 | | Crystal structure of monoclonal anti-CD4 antibody Q425 | | Descriptor: | Q425 Fab Light chain, Q425 Fab heavy chain | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6LKL
 
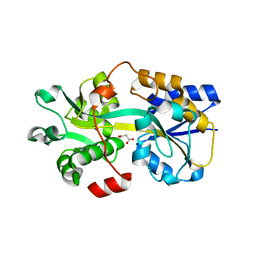 | | Two-component system protein mediate signal transduction | | Descriptor: | ABC transporter, solute-binding protein, MALONIC ACID | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKK
 
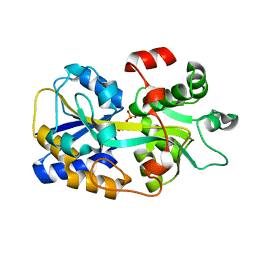 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKI
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | ABC transporter, solute-binding protein, MALONIC ACID, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKJ
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKG
 
 | | two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKH
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4QTS
 
 | |
8CEF
 
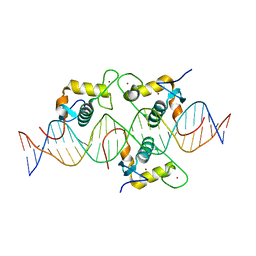 | | Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA | | Descriptor: | DNA (26-MER), Nuclear receptor DNA binding domain, ZINC ION | | Authors: | Patel, A.K.M, Shaik, T.B, McEwen, A.G, Moras, D, Klaholz, B.P, Billas, I.M.L. | | Deposit date: | 2023-02-01 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Nucleic Acids Res., 51, 2023
|
|
7SHP
 
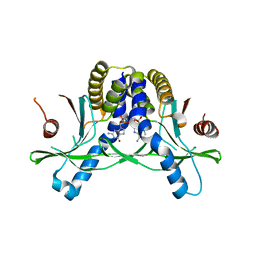 | | Crystal structure of hSTING in complex with c[2',3'-(ribo-2'-G, xylo-3'-A)-MP](RJ244) | | Descriptor: | (2S,5R,7R,8R,10S,12aR,14R,15R,15aR,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
7SHO
 
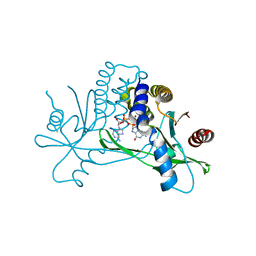 | | Crystal structure of hSTING in complex with c[2',3'-(ara-2'-G, ribo-3'-A)-MP] (RJ242) | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-10 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
3CXH
 
 | |
