2HIX
 
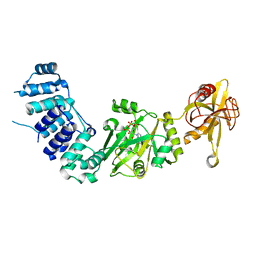 | |
2HIY
 
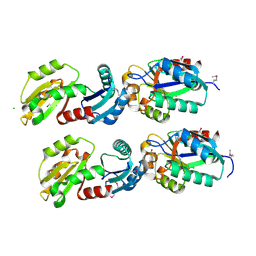 | | The structure of conserved bacterial protein SP0830 from Streptococcus pneumoniae. (CASP Target) | | Descriptor: | CHLORIDE ION, GLYCEROL, Hypothetical protein, ... | | Authors: | Cuff, M.E, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of conserved bacterial protein SP0830 from Streptococcus pneumoniae. (CASP Target)
To be Published
|
|
2HIZ
 
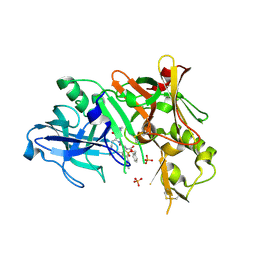 | | Crystal Structure of human beta-secretase (BACE) in the presence of an inhibitor | | Descriptor: | BENZYL [(1S)-2-({(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}AMINO)-2-OXO-1-{[(1-PROPYLBUTYL)SULFONYL]METHYL}ETHYL]CARBAMATE, Beta-secretase 1, PHOSPHATE ION | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-01-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of potent inhibitors of human beta-secretase. Part 1.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2HJ0
 
 | | Crystal Structure of the Putative Alfa Subunit of Citrate Lyase in Complex with Citrate from Streptococcus mutans, Northeast Structural Genomics Target SmR12 . | | Descriptor: | CITRIC ACID, Putative citrate lyase, alfa subunit | | Authors: | Forouhar, F, Hussain, M, Jayaraman, S, Shastry, R, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Alfa Subunit of Citrate Lyase in Complex with Citrate from Streptococcus mutans, Northeast Structural Genomics Target SmR12 (CASP Target).
To be Published
|
|
2HJ1
 
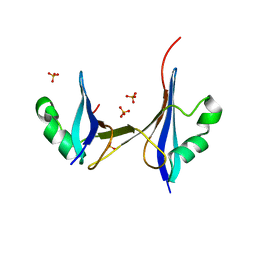 | | Crystal structure of a 3D domain-swapped dimer of protein HI0395 from Haemophilus influenzae | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y.V, Toro, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a 3D domain-swapped dimer of hypothetical protein from Haemophilus influenzae (CASP Target)
To be Published
|
|
2HJ3
 
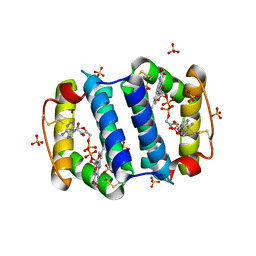 | | Structure of the Arabidopsis Thaliana Erv1 Thiol Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Sulfhydryl oxidase Erv1p | | Authors: | Vitu, E, Fass, D. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gain of Function in an ERV/ALR Sulfhydryl Oxidase by Molecular Engineering of the Shuttle Disulfide.
J.Mol.Biol., 362, 2006
|
|
2HJ4
 
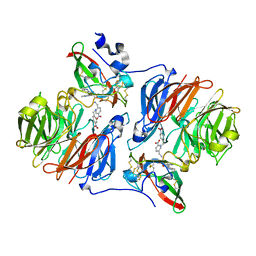 | |
2HJ6
 
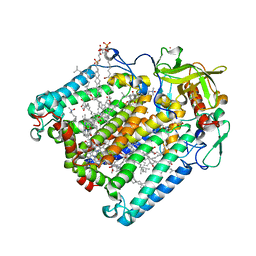 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylserine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-30 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HJ8
 
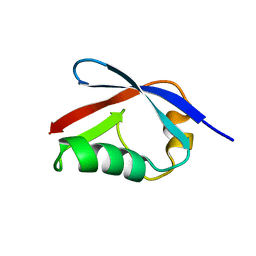 | | Solution NMR structure of the C-terminal domain of the interferon alpha-inducible ISG15 protein from Homo sapiens. Northeast Structural Genomics target HR2873B | | Descriptor: | Interferon-induced 17 kDa protein | | Authors: | Aramini, J.M, Ho, C.K, Yin, C, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal domain of the interferon alpha-inducible ISG15 protein from Homo sapiens. Northeast Structural Genomics target HR2873B.
To be Published
|
|
2HJ9
 
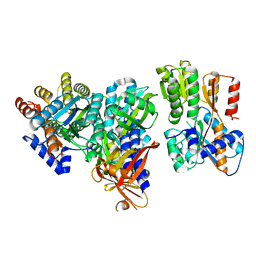 | | Crystal structure of the Autoinducer-2-bound form of Vibrio harveyi LuxP complexed with the periplasmic domain of LuxQ | | Descriptor: | 3A-METHYL-5,6-DIHYDRO-FURO[2,3-D][1,3,2]DIOXABOROLE-2,2,6,6A-TETRAOL, Autoinducer 2 sensor kinase/phosphatase luxQ, Autoinducer 2-binding periplasmic protein luxP | | Authors: | Neiditch, M.B, Hughson, F.M. | | Deposit date: | 2006-06-30 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing.
Cell(Cambridge,Mass.), 126, 2006
|
|
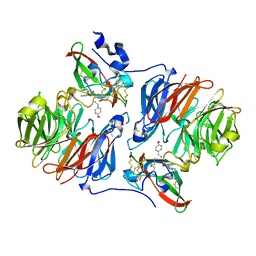
2HJB
 
 | |
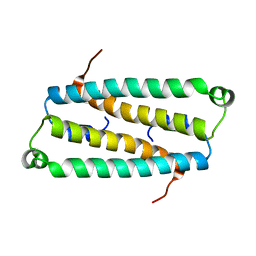
2HJD
 
 | |
2HJE
 
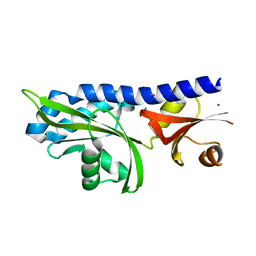 | |
2HJF
 
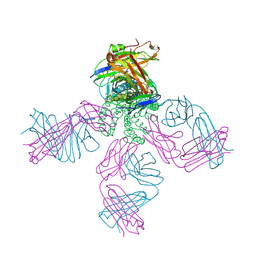 | | Potassium channel kcsa-fab complex with tetrabutylammonium (TBA) | | Descriptor: | Antibody fragment Heavy chain, Antibody fragment Light chain, POTASSIUM ION, ... | | Authors: | Faraldo-Gomez, J.D, Kutluay, E, Jogini, V, Zhao, Y, Heginbotham, L, Roux, B. | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Intracellular Block of the KcsA K(+) Channel by Tetrabutylammonium: Insights from X-ray Crystallography, Electrophysiology and Replica-exchange Molecular Dynamics Simulations.
J.Mol.Biol., 365, 2007
|
|
2HJG
 
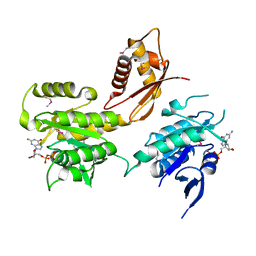 | | The crystal structure of the B. subtilis YphC GTPase in complex with GDP | | Descriptor: | GTP-binding protein engA, GUANOSINE-5'-DIPHOSPHATE, ZINC ION | | Authors: | Muench, S.P, Xu, L, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HJH
 
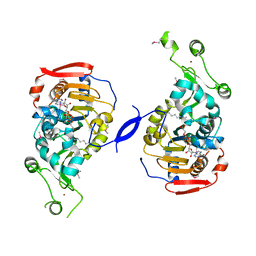 | | Crystal Structure of the Sir2 deacetylase | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, NAD-dependent histone deacetylase SIR2, NICOTINAMIDE, ... | | Authors: | Hall, B.E, Ellenberger, T.E. | | Deposit date: | 2006-06-30 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Autoregulation of the yeast Sir2 deacetylase by reaction and trapping of a pseudosubstrate motif in the active site
To be Published
|
|
2HJI
 
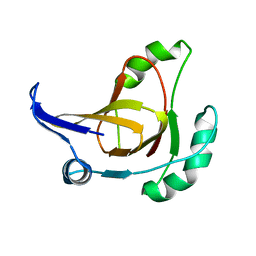 | | Structural model for the Fe-containing isoform of acireductone dioxygenase | | Descriptor: | E-2/E-2' protein, FE (II) ION | | Authors: | Pochapsky, T.C, Ju, T, Maroney, M.J, Chai, S.C. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | One Protein, Two Enzymes Revisited: A Structural Entropy Switch Interconverts the Two Isoforms of Acireductone Dioxygenase
J.Mol.Biol., 363, 2006
|
|
2HJJ
 
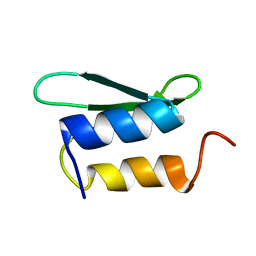 | | Solution NMR structure of protein ykfF from Escherichia coli. Northeast Structural Genomics target ER397. | | Descriptor: | Hypothetical protein ykfF | | Authors: | Swapna, G.V.T, Aramini, J.M, Zhao, L, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Liu, J, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein ykfF from Escherichia coli. Northeast Structural Genomics target ER397.
To be Published
|
|
2HJK
 
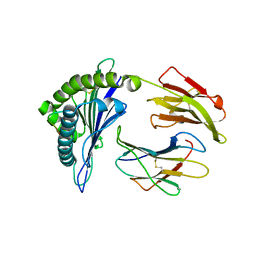 | | Crystal Structure of HLA-B5703 and HIV-1 peptide | | Descriptor: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen B-57 | | Authors: | Gillespie, G.M.A, Stewart-Jones, G, Rengasamy, J, Beattie, T, Bwayo, J.J, Plummer, F.A. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Strong TCR Conservation and Altered T Cell Cross-Reactivity Characterize a B*57-Restricted Immune Response in HIV-1 Infection.
J.Immunol., 177, 2006
|
|
2HJL
 
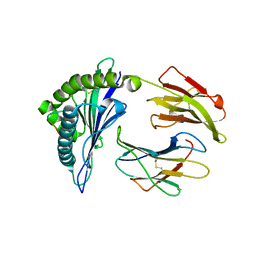 | | Crystal Structure of HLA-B5703 and HIV-1 peptide | | Descriptor: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen B-57 | | Authors: | Gillespie, G.M.A, Stewart-Jones, G, Rengasamy, J, Beattie, T, Bwayo, J.J, Plummer, F.A. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Strong TCR Conservation and Altered T Cell Cross-Reactivity Characterize a B*57-Restricted Immune Response in HIV-1 Infection.
J.Immunol., 177, 2006
|
|
2HJM
 
 | |
2HJN
 
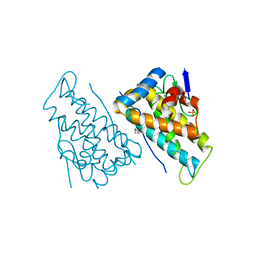 | |
2HJO
 
 | | Crystal structure of V224H design intermediate for GFP metal ion reporter | | Descriptor: | 1,2-ETHANEDIOL, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tubbs, J.L, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-06-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Iterative Structure-Based Design of a Green
Fluorescent Protein Metal Ion Reporter
To be Published
|
|
2HJP
 
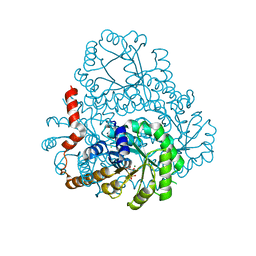 | |
2HJQ
 
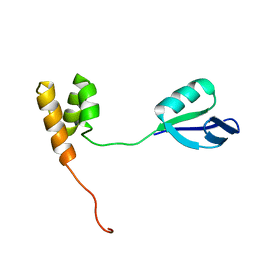 | | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449 | | Descriptor: | Hypothetical protein yqbF | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449
TO BE PUBLISHED
|
|
