3P9I
 
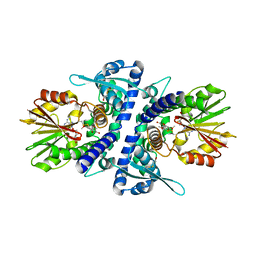 | | Crystal structure of perennial ryegrass LpOMT1 complexed with S-adenosyl-L-homocysteine and sinapaldehyde | | Descriptor: | (2E)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-enal, BETA-MERCAPTOETHANOL, Caffeic acid O-methyltransferase, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
3OYL
 
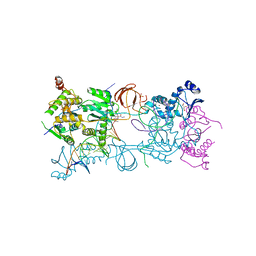 | | Crystal structure of the PFV S217H mutant intasome bound to magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYJ
 
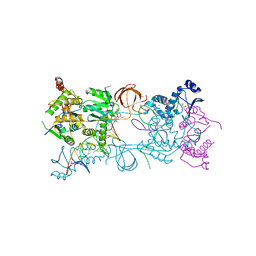 | | Crystal structure of the PFV S217Q mutant intasome in complex with magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
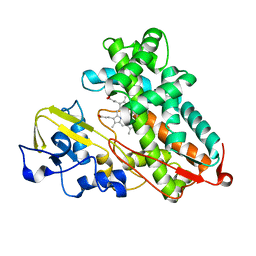
3P6R
 
 | |
3P6Q
 
 | |
3P80
 
 | |
3P90
 
 | | Crystal Structure Analysis of H207F Mutant of Human CLIC1 | | Descriptor: | Chloride intracellular channel protein 1 | | Authors: | Cross, M.O, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Dirr, H.W. | | Deposit date: | 2010-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of individual histidines in the pH-dependent global stability of human chloride intracellular channel 1.
Biochemistry, 51, 2012
|
|
3P9C
 
 | | Crystal structure of perennial ryegrass LpOMT1 bound to SAH | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
2AB1
 
 | | X-Ray Structure of Gene Product from Homo Sapiens HS.95870 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-26 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-Ray Structure of Gene Product from Homo Sapiens HS.95870
To be Published
|
|
3OPZ
 
 | | Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, Trans-sialidase, ... | | Authors: | Larrieux, N, Muia, R, Campetella, O, Buschiazzo, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-09 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Plos Pathog., 8, 2012
|
|
3LA6
 
 | | Octameric kinase domain of the E. coli tyrosine kinase Wzc with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Tyrosine-protein kinase wzc | | Authors: | Gruszczyk, J, Nessler, S, Gueguen-Chaignon, V, Vigouroux, A, Bechet, E, Grangeasse, C. | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export
Mol.Microbiol., 77, 2010
|
|
3LI2
 
 | | Closed Conformation of HtsA Complexed with Staphyloferrin A | | Descriptor: | (2R)-2-(2-{[(1R)-1-carboxy-4-{[(3S)-3,4-dicarboxy-3-hydroxybutanoyl]amino}butyl]amino}-2-oxoethyl)-2-hydroxybutanedioic acid, ACETATE ION, FE (III) ION, ... | | Authors: | Grigg, J.C, Murphy, M.E.P. | | Deposit date: | 2010-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket.
J.Biol.Chem., 285, 2010
|
|
3LMG
 
 | | Crystal structure of the ERBB3 kinase domain in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Shi, F, Lemmon, M.A. | | Deposit date: | 2010-01-30 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYE
 
 | |
3P6W
 
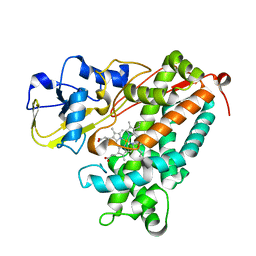 | |
3OYM
 
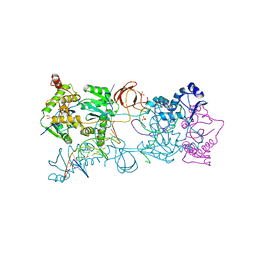 | | Crystal structure of the PFV N224H mutant intasome bound to manganese | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYH
 
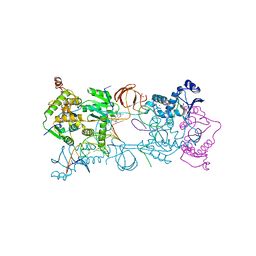 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI MK0536 | | Descriptor: | 6-(3-chloro-4-fluorobenzyl)-4-hydroxy-N,N-dimethyl-2-(1-methylethyl)-3,5-dioxo-2,3,5,6,7,8-hexahydro-2,6-naphthyridine-1-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3P6O
 
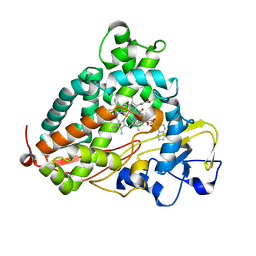 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-Etg-Dans | | Descriptor: | (3S,5S,7S)-N-(2-{2-[2-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)ethoxy]ethoxy}ethyl)tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-Etg-Dans
To be Published
|
|
3P8W
 
 | |
7FR6
 
 | |
2A7K
 
 | | carboxymethylproline synthase (CarB) from pectobacterium carotovora, apo enzyme | | Descriptor: | CarB | | Authors: | Sleeman, M.C, Sorensen, J.L, Batchelar, E.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2005-07-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Mechanistic Studies on Carboxymethylproline Synthase (CarB), a Unique Member of the Crotonase Superfamily Catalyzing the First Step in Carbapenem Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
7FR5
 
 | |
7FR2
 
 | |
7FRA
 
 | |
7FR0
 
 | |
