1D16
 
 | |
1D17
 
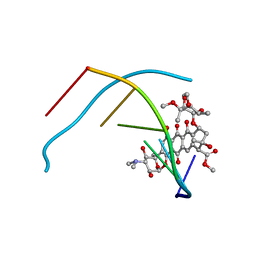 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
1D18
 
 | |
1D19
 
 | |
1D1A
 
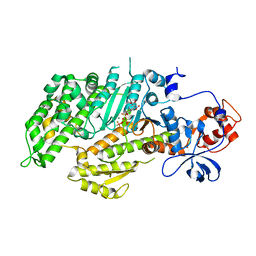 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O,P-DINITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, O,P-DINITROPHENYL AMINOETHYLDIPHOSPHATE-BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D1B
 
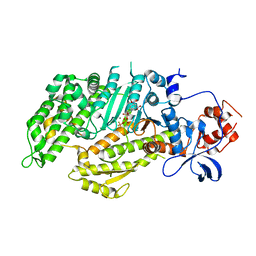 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O,P-DINITROPHENYL AMINOPROPYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, O,P-DINITROPHENYL AMINOPROPYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D1C
 
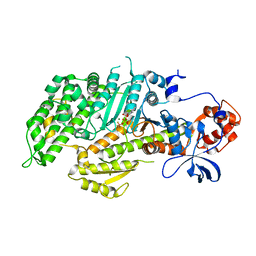 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH N-METHYL-O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, N-METHYL O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D1D
 
 | |
1D1E
 
 | |
1D1F
 
 | |
1D1G
 
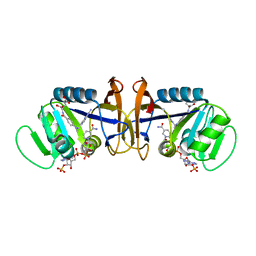 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-16 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
1D1H
 
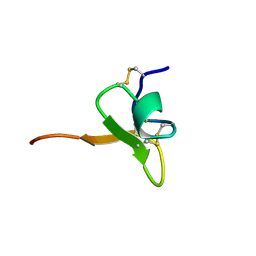 | | SOLUTION STRUCTURE OF HANATOXIN 1 | | Descriptor: | HANATOXIN TYPE 1 | | Authors: | Takahashi, H, Kim, J.I, Sato, K, Swartz, K.J, Shimada, I. | | Deposit date: | 1999-09-16 | | Release date: | 2000-09-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hanatoxin1, a gating modifier of voltage-dependent K(+) channels: common surface features of gating modifier toxins.
J.Mol.Biol., 297, 2000
|
|
1D1I
 
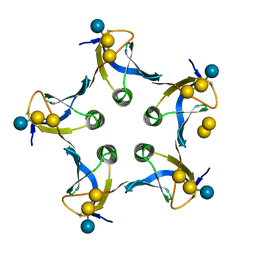 | |
1D1J
 
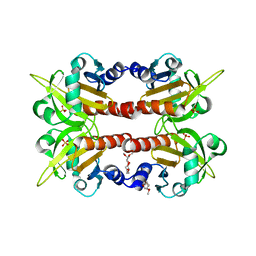 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
1D1K
 
 | |
1D1L
 
 | | CRYSTAL STRUCTURE OF CRO-F58W MUTANT | | Descriptor: | LAMBDA CRO REPRESSOR, SULFATE ION | | Authors: | Rupert, P.B, Mollah, A.K, Mossing, M.C, Matthews, B.W. | | Deposit date: | 1999-09-17 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for enhanced stability and reduced DNA binding seen in engineered second-generation Cro monomers and dimers.
J.Mol.Biol., 296, 2000
|
|
1D1M
 
 | |
1D1N
 
 | | SOLUTION STRUCTURE OF THE FMET-TRNAFMET BINDING DOMAIN OF BECILLUS STEAROTHERMOPHILLUS TRANSLATION INITIATION FACTOR IF2 | | Descriptor: | INITIATION FACTOR 2 | | Authors: | Meunier, S, Spurio, S, Czisch, M, Wechselberger, R, Gueunneugues, M. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2.
EMBO J., 19, 2000
|
|
1D1O
 
 | |
1D1P
 
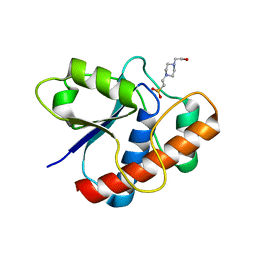 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1D1Q
 
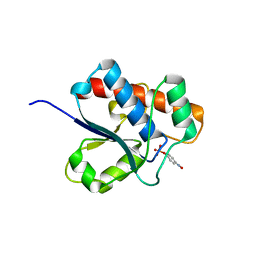 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) COMPLEXED WITH THE SUBSTRATE PNPP | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Staufacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1D1R
 
 | |
1D1S
 
 | | WILD-TYPE HUMAN SIGMA (CLASS IV) ALCOHOL DEHYDROGENASE | | Descriptor: | ACETATE ION, ALCOHOL DEHYDROGENASE CLASS IV SIGMA CHAIN, CACODYLATE ION, ... | | Authors: | Xie, P.T, Hurley, T.D. | | Deposit date: | 1999-09-21 | | Release date: | 1999-09-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Methionine-141 directly influences the binding of 4-methylpyrazole in human sigma sigma alcohol dehydrogenase.
Protein Sci., 8, 1999
|
|
1D1T
 
 | |
1D1U
 
 | | USE OF AN N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE TO FACILITATE CRYSTALLIZATION AND ANALYSIS OF A PSEUDO-16-MER DNA MOLECULE CONTAINING G-A MISPAIRS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*GP*TP*G)-3'), PROTEIN (REVERSE TRANSCRIPTASE) | | Authors: | Cote, M.L, Yohannan, S, Georgiadis, M.M. | | Deposit date: | 1999-09-21 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
