2JXP
 
 | | Solution NMR structure of uncharacterized lipoprotein B from Nitrosomonas europaea. Northeast Structural Genomics target NeR45A | | Descriptor: | Putative lipoprotein B | | Authors: | Rossi, P, Wang, D, Janjua, H, Owens, L, Xiao, R, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of uncharacterized lipoprotein B from Nitrosomonas europaea.
To be Published
|
|
1JSX
 
 | |
4FN5
 
 | |
1XH7
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-[4-({4-[5-(3,3-DIMETHYLPIPERIDIN-1-YL)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH4
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-[4-({4-[5-(DIMETHYLAMINO)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1YHT
 
 | | Crystal structure analysis of Dispersin B | | Descriptor: | ACETIC ACID, DspB, GLYCEROL | | Authors: | Ramasubbu, N, Thomas, L.M, Ragunath, C, Kaplan, J.B. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Dispersin B, a Biofilm-releasing Glycoside Hydrolase from the Periodontopathogen Actinobacillus actinomycetemcomitans.
J.Mol.Biol., 349, 2005
|
|
2GPB
 
 | |
3BP9
 
 | | Structure of B-tropic MLV capsid N-terminal domain | | Descriptor: | GLYCEROL, Gag protein, ISOPROPYL ALCOHOL | | Authors: | Gulnahar, M.B, Dodding, M.P, Goldstone, D.C, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation
J.Mol.Biol., 376, 2008
|
|
1AQ7
 
 | |
1C3B
 
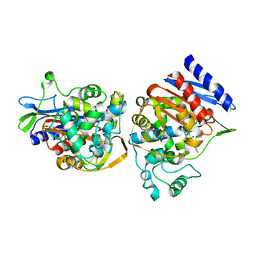 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR, BENZO(B)THIOPHENE-2-BORONIC ACID (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, CEPHALOSPORINASE | | Authors: | Powers, R.A, Blazquez, J, Weston, G.S, Morosini, M.I, Baquero, F, Shoichet, B.K. | | Deposit date: | 1999-07-27 | | Release date: | 1999-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The complexed structure and antimicrobial activity of a non-beta-lactam inhibitor of AmpC beta-lactamase.
Protein Sci., 8, 1999
|
|
1XH9
 
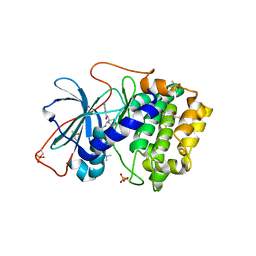 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-[4-({4-[5-(DIMETHYLAMINO)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1XH5
 
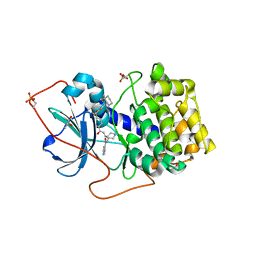 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-{4-[(4-{3-[(2R)-3,3-DIMETHYLPIPERIDIN-2-YL]-2-FLUORO-6-HYDROXYBENZOYL}BENZOYL)AMINO]AZEPAN-3-YL}ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
4J6K
 
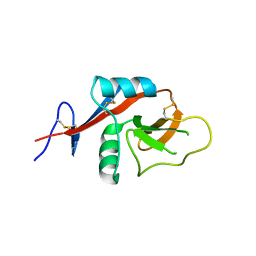 | |
1ZYS
 
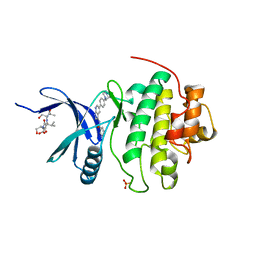 | | Co-crystal structure of Checkpoint Kinase Chk1 with a pyrrolo-pyridine inhibitor | | Descriptor: | N-{5-[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]-1H-PYRROLO[2,3-B]PYRIDIN-3-YL}NICOTINAMIDE, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Stavenger, R.A, Zhao, B, Zhou, B.-B.S, Brown, M.J, Lee, D, Holt, D.A. | | Deposit date: | 2005-06-10 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolo[2,3-b]pyridines Inhibit the Checkpoint Kinase Chk1
To be Published
|
|
3EX9
 
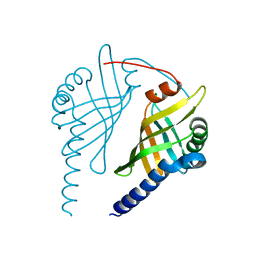 | |
3CVK
 
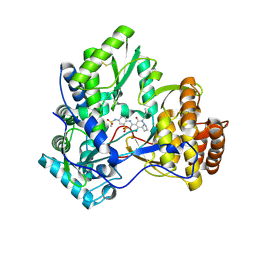 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[1-(3,3-Dimethyl-butyl)-4-hydroxy-2-oxo-1,2,4a,5,6,7-hexahydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,2-dihydro -1lambda6-benzo[1,2,4]thiadiazin-7-yl}-methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-04-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hexahydro-pyrrolo- and hexahydro-1H-pyrido[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4IWD
 
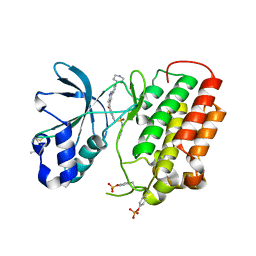 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | Descriptor: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
1DMM
 
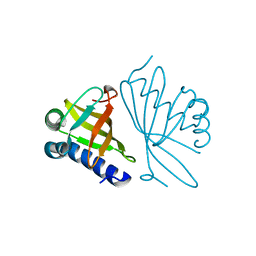 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
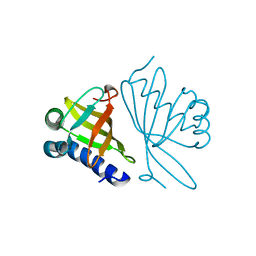 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMQ
 
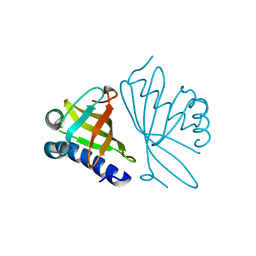 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
3CNM
 
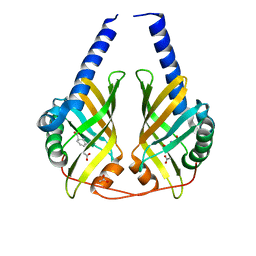 | | Crystal Structure of Phenazine Biosynthesis Protein PhzA/B from Burkholderia cepacia R18194, DHHA complex | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3DZL
 
 | | Crystal structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-3-oxocyclohexanecarboxylic acid | | Descriptor: | (1R)-3-oxocyclohexanecarboxylic acid, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2008-07-30 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PhzA/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
159D
 
 | | SIDE BY SIDE BINDING OF TWO DISTAMYCIN A DRUGS IN THE MINOR GROOVE OF AN ALTERNATING B-DNA DUPLEX | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*IP*CP*IP*CP*IP*CP*IP*C)-3'), MAGNESIUM ION | | Authors: | Chen, X, Ramakrishnan, B, Rao, S.T, Sundaralingam, M. | | Deposit date: | 1994-02-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Nat.Struct.Biol., 1, 1994
|
|
4GGL
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | Descriptor: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)pyrido[2,3-b]pyrazin-2(1H)-one, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-08-06 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4GPB
 
 | |
