1UIU
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel unliganded form) | | Descriptor: | Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIV
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel liganded form) | | Descriptor: | NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
5ZBU
 
 | |
1R0C
 
 | |
2LFE
 
 | | Solution NMR structure of N-terminal domain of human E3 ubiquitin-protein ligase HECW2, Northeast structural genomics consortium (NESG) target ht6306A | | Descriptor: | E3 ubiquitin-protein ligase HECW2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Chitayat, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-29 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of E3 ligase HECW2
To be Published
|
|
2WQW
 
 | |
4E1G
 
 | |
5IJ6
 
 | | Crystal structure of Enterococcus faecalis lipoate-protein ligase A (lplA-1) in complex with lipoic acid | | Descriptor: | CHLORIDE ION, LIPOIC ACID, Lipoate--protein ligase, ... | | Authors: | Hughes, S.J, Lyle, A.G, Song, J.H, Antoshchenko, T, Park, H. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enterococcus faecalis lipoate-protein ligase A (lplA-1) in complex with lipoic acid
to be published
|
|
2AN6
 
 | | Protein-peptide complex | | Descriptor: | Ubiquitin ligase SIAH1A, ZINC ION, peptide from Phyllopod | | Authors: | House, C.M, Hancock, N.C, Moller, A, Cromer, B.A, Fedorov, V, Bowtell, D.D.L, Parker, M.W, Polekhina, G. | | Deposit date: | 2005-08-11 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidation of the substrate binding site of Siah ubiquitin ligase
Structure, 14, 2006
|
|
4GD0
 
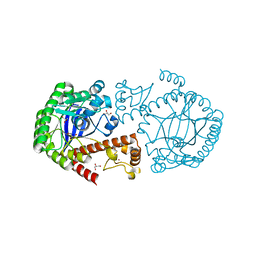 | | tRNA-guanine transglycosylase Y106F, C158V mutant | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Schmidt, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
3LRQ
 
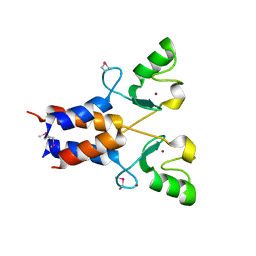 | | Crystal structure of the U-box domain of human ubiquitin-protein ligase (E3), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D. | | Descriptor: | E3 ubiquitin-protein ligase TRIM37, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | E3 UBIQUITIN-PROTEIN LIGASE RING DOMAIN FROM HUMAN TRIPARTITE MOTIF-CONTAINING PROTEIN 37 (TRIM37), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D.
To be Published
|
|
3LP8
 
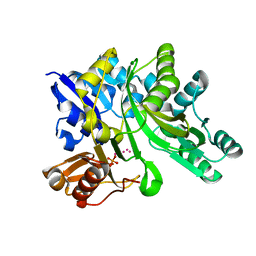 | |
6XXW
 
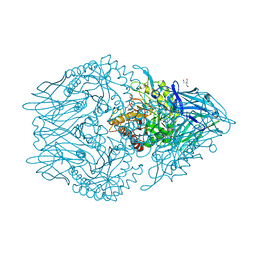 | | Structure of beta-D-Glucuronidase for Dictyoglomus thermophilum. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucuronidase, ... | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Thioglycoligation of aromatic thiols using a natural glucuronide donor.
Org.Biomol.Chem., 18, 2020
|
|
2WQU
 
 | |
1X4S
 
 | | Solution structure of zinc finger HIT domain in protein FON | | Descriptor: | ZINC ION, Zinc finger HIT domain containing protein 2 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger HIT domain in protein FON
Protein Sci., 16, 2007
|
|
3Q12
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis in complex with pantoate. | | Descriptor: | CHLORIDE ION, PANTOATE, Pantoate--beta-alanine ligase | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
6BLJ
 
 | |
1UKZ
 
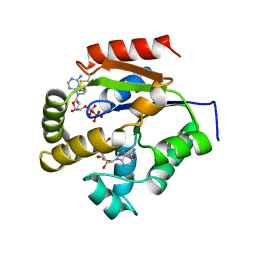 | |
4CI2
 
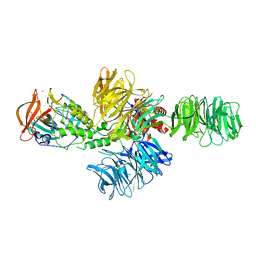 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
1UKY
 
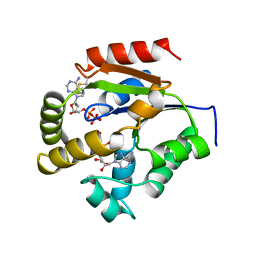 | |
5ICL
 
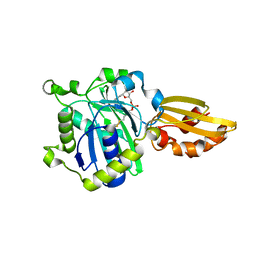 | |
1YIU
 
 | | Itch E3 ubiquitin ligase WW3 domain | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Shaw, A.Z, Martin-Malpartida, P, Morales, B, Yraola, F, Royo, M, Macias, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of either Ser16 or Thr30 does not disrupt the structure of the Itch E3 ubiquitin ligase third WW domain
Proteins, 60, 2005
|
|
4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
3MVN
 
 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
6BRQ
 
 | | Crystal structure of rice ASK1-D3 ubiquitin ligase complex crystal form 3 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
