8YLA
 
 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9BEI
 
 | |
6Z15
 
 | | Human wtSTING in complex with 3',3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z0Z
 
 | | Human wtSTING in complex with 3',3'-c-(2'FdAMP-2'FdAMP) | | Descriptor: | 2'-fluoro-,3',3'-c-di-AMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7R4H
 
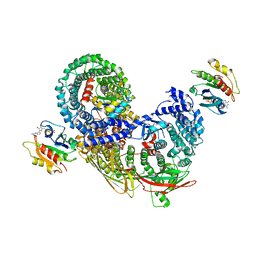 | | phospho-STING binding to adaptor protein complex-1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Xu, P, Ablasser, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Clathrin-associated AP-1 controls termination of STING signalling.
Nature, 610, 2022
|
|
7JFS
 
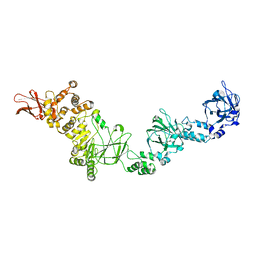 | |
6XNN
 
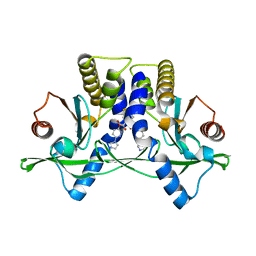 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6L7Z
 
 | | Solution NMR structure of the N-terminal immunoglobulin variable domain of BTNL2 | | Descriptor: | Butyrophilin-like protein 2 | | Authors: | Basak, A.J, Lee, W, Samanta, D, De, S. | | Deposit date: | 2019-11-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into N-terminal IgV Domain of BTNL2, a T Cell Inhibitory Molecule, Suggests a Non-canonical Binding Interface for Its Putative Receptors.
J.Mol.Biol., 432, 2020
|
|
5L36
 
 | |
7JNB
 
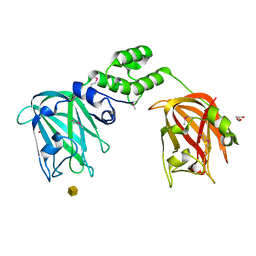 | |
7JS4
 
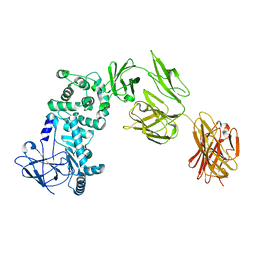 | |
7ZCJ
 
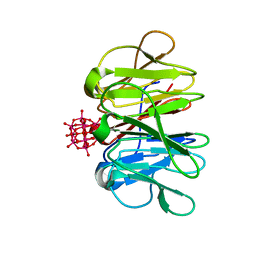 | | Crystal structure of Pizza6-TNH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
6AKE
 
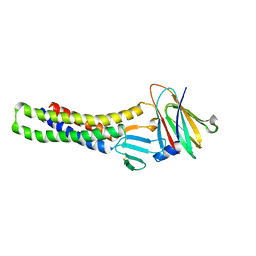 | |
6H7B
 
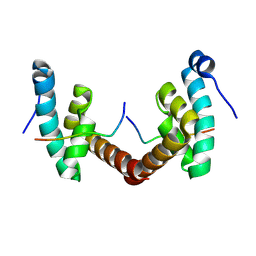 | | Structure of Leishmania PABP1 (domain J) complexed with a peptide containing the PAM2 motif of eIF4E4. | | Descriptor: | HIS-HIS-MET-ASN-PRO-ASN-ALA-THR-GLU-PHE-MET-PRO, Polyadenylate-binding protein | | Authors: | Cameron, A.D, Firczuk, H, dos Santos Rodrigues, F.H, McCarthy, J.E.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Leishmania PABP1-eIF4E4 interface: a novel 5'-3' interaction architecture for trans-spliced mRNAs.
Nucleic Acids Res., 47, 2019
|
|
6MGA
 
 | |
6S9K
 
 | | Structure of 14-3-3 gamma in complex with caspase-2 peptide containing 14-3-3 binding motif Ser139 and NLS | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, Caspase-2 | | Authors: | Alblova, M, Obsil, T, Obsilova, V. | | Deposit date: | 2019-07-15 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 14-3-3 protein binding blocks the dimerization interface of caspase-2.
Febs J., 287, 2020
|
|
6SAD
 
 | |
7SSM
 
 | | Crystal structure of human STING R232 in complex with compound 11 | | Descriptor: | 2-({[(8R)-pyrazolo[1,5-a]pyrimidine-3-carbonyl]amino}methyl)-1-benzofuran-7-carboxylic acid, Stimulator of interferon genes protein | | Authors: | Sack, J.S, Critton, D.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Non-Nucleotide Small-Molecule STING Agonists via Chemotype Hybridization.
J.Med.Chem., 65, 2022
|
|
8A2K
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-[(2-naphthyloxy)methyl]phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-[4-(naphthalen-1-yloxymethyl)phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2H
 
 | | human STING in complex with 2',3'-cyclic-GMP-7-deazaphenyl-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanyl-5-phenyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Smola, M, Vavrina, Z, Boura, E, Brynda, J. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2J
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-biphenylyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-phenylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2I
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-(2-naphthyl)phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-naphthalen-2-ylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
6H6S
 
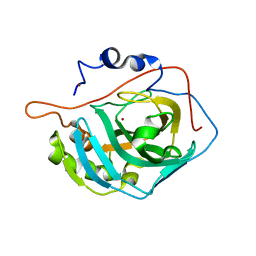 | | Sad phasing on nickel-substituted human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, NICKEL (II) ION | | Authors: | Calderone, V, Fragai, M, Silva, J.P, Luchinat, C, Ravera, E, Geraldes, C.F.G.C, Macedo, A.L, Cerofolini, L, Giuntini, S. | | Deposit date: | 2018-07-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-crystallographic symmetry in proteins: Jahn-Teller-like and Butterfly-like effects?
J. Biol. Inorg. Chem., 24, 2019
|
|
8A0D
 
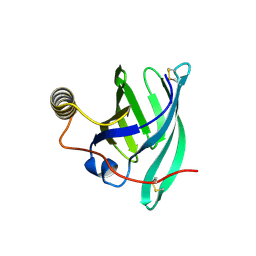 | | Crystal structure of the major guinea pig allergen Cav p 1.0101 part of the lipocalin family | | Descriptor: | Allergen lipocalin Cav p 1 isoform 1 | | Authors: | Herman, R, Charlier, P, Janssen-Weets, B, Hilger, C, Swiontek, K. | | Deposit date: | 2022-05-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.685 Å) | | Cite: | Mammalian derived lipocalin and secretoglobin respiratory allergens strongly bind ligands with potentially immune modulating properties.
Front Allergy, 3, 2022
|
|
5IXD
 
 | |
