3U9S
 
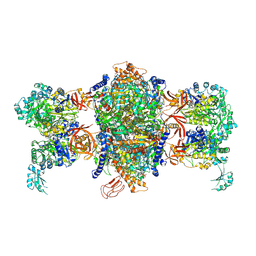 | | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Methylcrotonyl-CoA carboxylase, ... | | Authors: | Huang, C.S, Tong, L. | | Deposit date: | 2011-10-19 | | Release date: | 2011-12-14 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An unanticipated architecture of the 750-kDa {alpha}6{beta}6 holoenzyme of 3-methylcrotonyl-CoA carboxylase
Nature, 481, 2012
|
|
4E2V
 
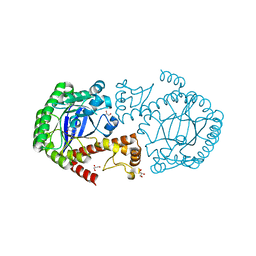 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with preQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Schmidt, I, Reuter, K, Klebe, G, Heine, A, Tidten, N. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
5M45
 
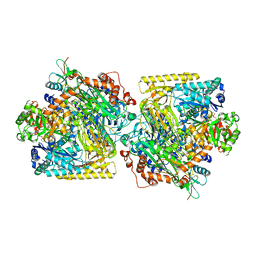 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
3TT6
 
 | |
5A2X
 
 | | Crystal structure of mtPAP in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
2XFY
 
 | | Crystal structure of Barley Beta-Amylase complexed with alpha- cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, BETA-AMYLASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
4E1V
 
 | | X-RAY Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with 5-Fluorouracil at 2.15 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Lashkov, A.A, Sotnichenko, S.E, Prokofev, I.I, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2012-03-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structure of Salmonella typhimurium uridine phosphorylase complexed with 5-fluorouracil and molecular modelling of the complex of 5-fluorouracil with uridine phosphorylase from Vibrio cholerae.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5LID
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromopromazine | | Descriptor: | Cys-loop ligand-gated ion channel, bromopromazine | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5AHI
 
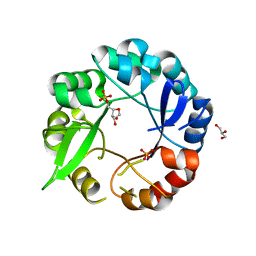 | | Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanism of Hisa from Salmonella Enterica
To be Published
|
|
4P73
 
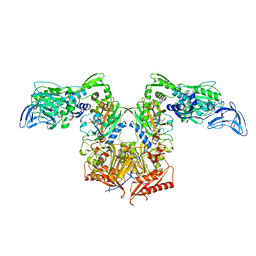 | | PheRS in complex with compound 1a | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
1Q0W
 
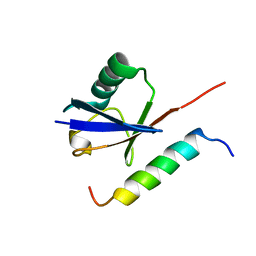 | | Solution structure of Vps27 amino-terminal UIM-ubiquitin complex | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS27 | | Authors: | Swanson, K.A, Kang, R.S, Stamenova, S.D, Hicke, L, Radhakrishnan, I. | | Deposit date: | 2003-07-17 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vps27 UIM-Ubiquitin Complex Important for Endosomal Sorting and Receptor Downregulation
Embo J., 22, 2003
|
|
4PLA
 
 | |
5LKV
 
 | |
1Q2D
 
 | |
5LL1
 
 | |
5AGK
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (2S)-2-Amino-5-(2-(methylsulfinyl)acetimidamido) pentanoic acid | | Descriptor: | (2S)-2-AMINO-5-(2-(METHYLSULFINYL)ACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
8RJA
 
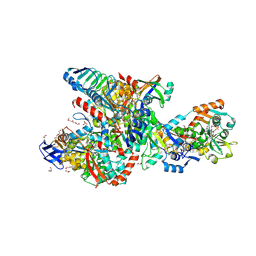 | | Crystal structure of the F420-reducing formylmethanofuran dehydrogenase complex from the ethanotroph Candidatus Ethanoperedens thermophilum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-12-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | F420 reduction as a cellular driver for anaerobic ethanotrophy
To Be Published
|
|
5AEA
 
 | |
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
5AFZ
 
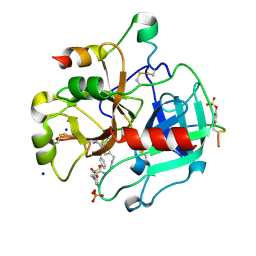 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-(2-((4- carbamimidoylphenyl)methylamino)-2-oxo-propyl)-3-phenyl-propanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, N-(BENZYLSULFONYL)-D-PHENYLALANYL-N-(4-CARBAMIMIDOYLBENZYL)GLYCINAMIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
3TYR
 
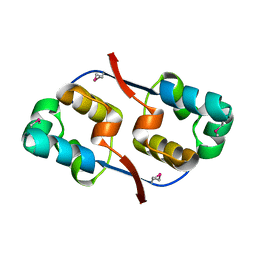 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
1PMB
 
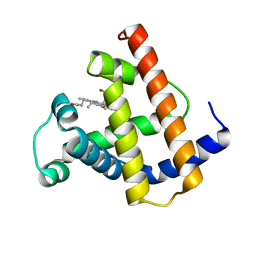 | | THE DETERMINATION OF THE CRYSTAL STRUCTURE OF RECOMBINANT PIG MYOGLOBIN BY MOLECULAR REPLACEMENT AND ITS REFINEMENT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Dodson, E.J, Dodson, G.G, Hubbard, R.E, Wilkinson, A.J. | | Deposit date: | 1989-11-27 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the crystal structure of recombinant pig myoglobin by molecular replacement and its refinement.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
2XFB
 
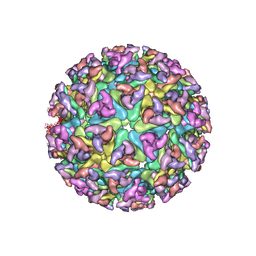 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SINDBIS VIRUS cryo- EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
8TVR
 
 | |
5A0L
 
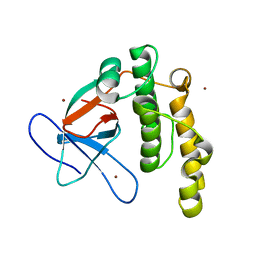 | | N-terminal thioester domain of fibronectin-binding protein SfbI from Streptococcus pyogenes | | Descriptor: | ACETATE ION, FIBRONECTIN-BINDING PROTEIN, ZINC ION | | Authors: | Walden, M, Edwards, J.M, Dziewulska, A.M, Kan, S.-Y, Schwarz-Linek, U, Banfield, M.J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An internal thioester in a pathogen surface protein mediates covalent host binding.
Elife, 4, 2015
|
|
