7CLL
 
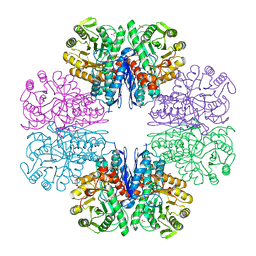 | | Mycobacterium tubeculosis enolase in complex with 2-Phosphoglycerate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
7CKP
 
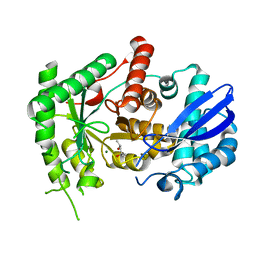 | | Mycobacterium tuberculosis Enolase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enolase, MAGNESIUM ION | | Authors: | Biswal, B.K, Ahmad, M, Jha, B. | | Deposit date: | 2020-07-18 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
2WJP
 
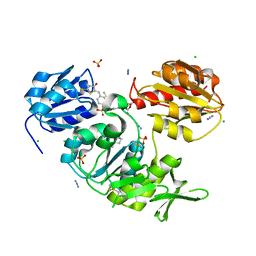 | | CRYSTAL STRUCTURE OF MURD LIGASE IN COMPLEX WITH D-GLU CONTAINING RHODANINE INHIBITOR | | Descriptor: | AZIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Rupnik, V, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Muller-Premru, M, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2009-05-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel 5-Benzylidenerhodanine and 5-Benzylidenethiazolidine-2,4-Dione Inhibitors of Murd Ligase.
J.Med.Chem., 53, 2010
|
|
7DLR
 
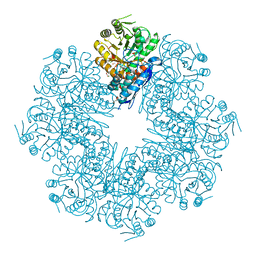 | | Mycobacterium tuberculosis enolase mutant - E163A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ahmad, M, Biswal, B.K. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
2XYV
 
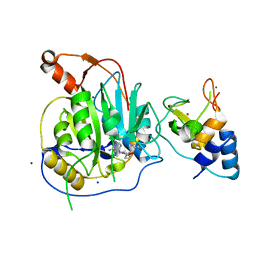 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYR
 
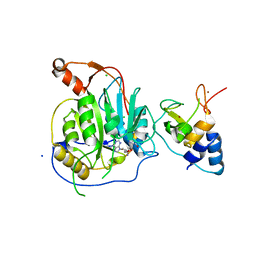 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
7CLK
 
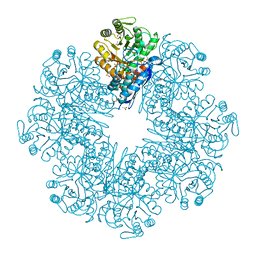 | | Mycobacterium tuberculosis enolase in complex with alternate 2-phosphoglycerate | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, ACETATE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-07-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
2XPC
 
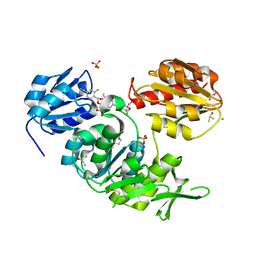 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
3HPE
 
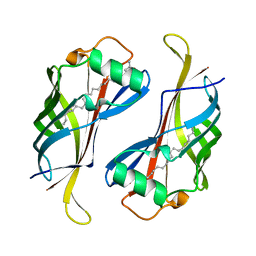 | |
2XYQ
 
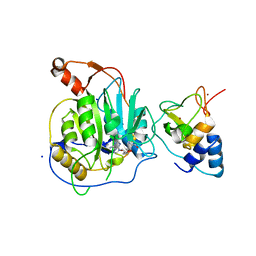 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2Y1O
 
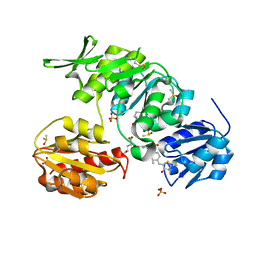 | | Dual-target Inhibitor of MurD and MurE Ligases: Design, Synthesis and Binding Mode Studies | | Descriptor: | (2R)-2-[[3-[[4-[(Z)-(4-OXO-2-SULFANYLIDENE-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL]METHYLAMINO]PHENYL]CARBONYLAMINO]PENTANEDIOIC ACID, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Tomasic, T, Sink, R, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Kikelj, D, Peterlin-Masic, L. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Dual Inhibitor of MurD and MurE Ligases from Escherichia coli and Staphylococcus aureus.
ACS Med Chem Lett, 3, 2012
|
|
2Y67
 
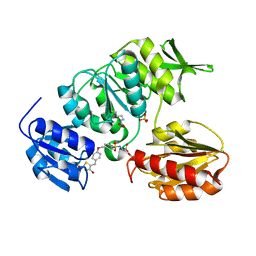 | | New 5-Benzylidenethiazolidine-4-one Inhibitors of Bacterial MurD Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation | | Descriptor: | (2R)-2-[[4-[[4-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]phenoxy]methyl]phenyl]sulfonylamino]pentanedioic acid, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Zidar, N, Tomasic, T, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 5-Benzylidenethiazolidin-4-One Inhibitors of Bacterial Murd Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation.
Eur.J.Med.Chem, 46, 2011
|
|
7E4F
 
 | | Mycobacterium tuberculosis enolase mutant - E204A complex with phosphoenolpyruvate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahmad, M, Pal, R.K, Biswal, B.K. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
3KVD
 
 | | Crystal structure of the Neisseria meningitidis Factor H binding protein, fHbp (GNA1870) at 2.0 A resolution | | Descriptor: | Lipoprotein | | Authors: | Cendron, L, Veggi, D, Girardi, E, Zanotti, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the uncomplexed Neisseria meningitidis factor H-binding protein fHbp (rLP2086).
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2Y68
 
 | | Structure-based design of a new series of D-glutamic acid-based inhibitors of bacterial MurD ligase | | Descriptor: | 2-[[2-fluoro-5-[[[4-[(Z)-(4-oxo-2-sulfanylidene-1,3-thiazolidin-5-ylidene)methyl]phenyl]amino]methyl]phenyl]carbonylamino]pentanedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-based design of a new series of D-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD).
J. Med. Chem., 54, 2011
|
|
3VFL
 
 | |
3ZXX
 
 | | Structure of self-cleaved protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
3ZXY
 
 | | Structure of S218A mutant of the protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
4AQF
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
4AQG
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
1ETE
 
 | |
2J0A
 
 | | Structure of the catalytic domain of mouse Manic Fringe | | Descriptor: | BETA-1,3-N-ACETYLGLUCOSAMINYLTRANSFERASE MANIC FRINGE, POTASSIUM ION, SULFATE ION | | Authors: | Jinek, M, Chen, Y.-W, Clausen, H, Cohen, S.M, Conti, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Notch-Modifying Glycosyltransferase Fringe
Nat.Struct.Mol.Biol., 13, 2006
|
|
2KVY
 
 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
2I3V
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
