6FPA
 
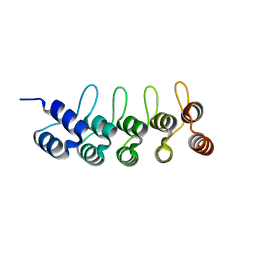 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
8E15
 
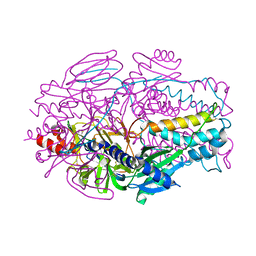 | | A computationally stabilized hMPV F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F1 protein with Fibritin peptide, F2 protein, ... | | Authors: | Huang, J, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Biorxiv, 2023
|
|
6G6N
 
 | | Crystal structure of the computationally designed Tako8 protein in C2 | | Descriptor: | Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G6M
 
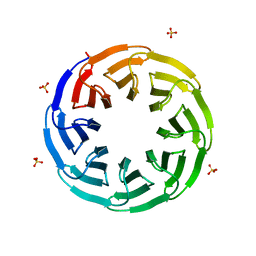 | | Crystal structure of the computationally designed Tako8 protein in P42212 | | Descriptor: | SULFATE ION, Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G6Q
 
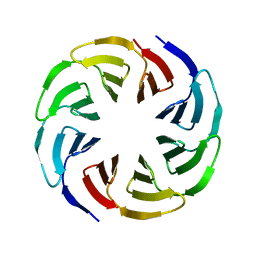 | | Crystal structure of the computationally designed Ika4 protein | | Descriptor: | Ika4 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6FPB
 
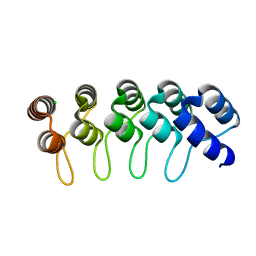 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group I4 | | Descriptor: | CHLORIDE ION, DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
4AFS
 
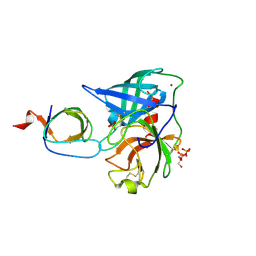 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, GLYCEROL, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
6C4Y
 
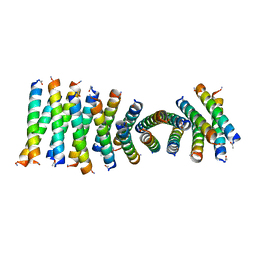 | | Cross-alpha Amyloid-like Structure alphaAmG | | Descriptor: | Cross-alpha Amyloid-like Structure alphaAmG | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
4AFZ
 
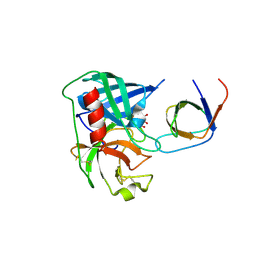 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, D(-)-TARTARIC ACID, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4A29
 
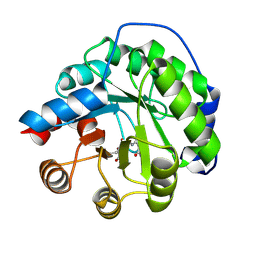 | | Structure of the engineered retro-aldolase RA95.0 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
6FCE
 
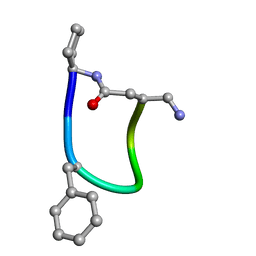 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-23 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
4AFQ
 
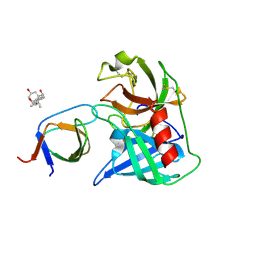 | | Human Chymase - Fynomer Complex | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHYMASE, CITRATE ANION, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
6NBR
 
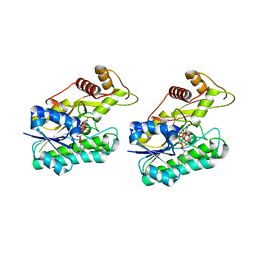 | |
6C4Z
 
 | |
4AG2
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHYMASE, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AG1
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, SULFATE ION | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
6D37
 
 | |
8OT1
 
 | |
8OT3
 
 | |
8OT4
 
 | |
6CFA
 
 | | peptide PaAMP1R3 | | Descriptor: | peptide PaAMP1R3 | | Authors: | Alves, E.S.F, Liao, L.M. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic peptide PaAMP1R3
To be Published
|
|
6AZF
 
 | |
6OP5
 
 | |
6G6O
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.1 in P63 | | Descriptor: | GLYCEROL, Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
8PEQ
 
 | | Complex of diubiquitin-derived artificial binding protein (Affilin) variant Af2 with its target oncofetal fibronectin (fragment 7B8) | | Descriptor: | Affilin variant Af2, Fibronectin, SULFATE ION | | Authors: | Parthier, C, Katzschmann, A, Fiedler, E, Haupts, U, Reimann, A. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Ubiquitin-derived artificial binding proteins targeting oncofetal fibronectin reveal scaffold plasticity by beta-strand slippage
to be published
|
|
