8B92
 
 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound SI-2) | | Descriptor: | 4-chloranyl-6-fluoranyl-~{N}3-[2-fluoranyl-4-(oxetan-3-yl)phenyl]-~{N}1-[(2-methoxyphenyl)methyl]benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B95
 
 | | Crystal structure of PPARG and NCOR2 with BAY-9683, an inverse agonist | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}1-[[3,4-bis(fluoranyl)phenyl]methyl]-4-chloranyl-6-fluoranyl-~{N}3-(3-methyl-5-morpholin-4-yl-pyridin-2-yl)benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B94
 
 | | Crystal structure of PPARG and NCOR2 with BAY-5516, an inverse agonist | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}3-[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]-4-chloranyl-6-fluoranyl-~{N}1-[(4-fluorophenyl)methyl]benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B8Z
 
 | | Crystal structure of mutant PPARG (C313A) and NCOR2 with an inverse agonist (compound 7e) | | Descriptor: | 4,5-bis(chloranyl)-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B8W
 
 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 7a) | | Descriptor: | 4-chloranyl-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, GLYCEROL, Nuclear receptor corepressor 2, ... | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B91
 
 | | Crystal structure of mutant PPARG (C313A) and NCOR2 with an inverse agonist (compound SI-1) | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}3-[4-[bis(fluoranyl)methoxy]-3-fluoranyl-phenyl]-4-chloranyl-6-fluoranyl-~{N}1-[(2-methoxyphenyl)methyl]benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B90
 
 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 7d) | | Descriptor: | 5-chloranyl-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
5JGJ
 
 | | Crystal structure of GtmA | | Descriptor: | UbiE/COQ5 family methyltransferase, putative | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
2H1M
 
 | |
3BXD
 
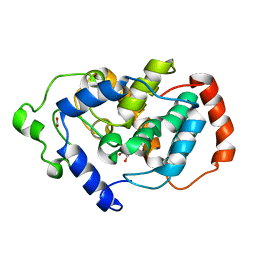 | | Crystal structure of Mouse Myo-inositol oxygenase (re-refined) | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Hallberg, B.M. | | Deposit date: | 2008-01-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterization of human myo-inositol oxygenase.
J.Biol.Chem., 283, 2008
|
|
5KCQ
 
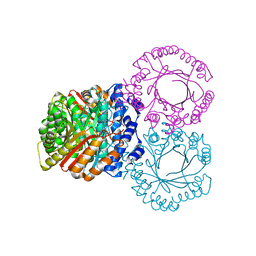 | |
5KDA
 
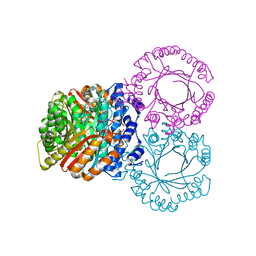 | |
5KCY
 
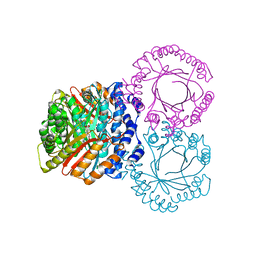 | |
5KCL
 
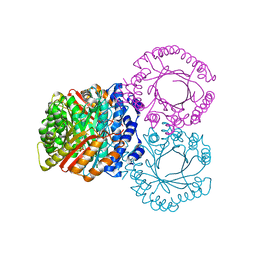 | |
5KD0
 
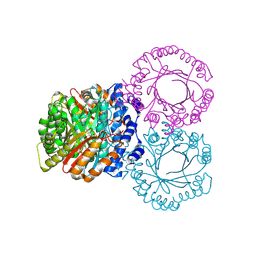 | |
6NCW
 
 | |
7ZKG
 
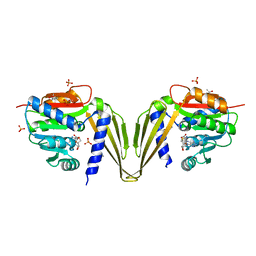 | | C-Methyltransferase PsmD from Streptomyces griseofuscus with bound cofactor (crystal form 2) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-13 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
7ZGT
 
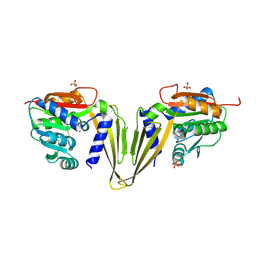 | | C-Methyltransferase PsmD from Streptomyces griseofuscus (apo form) | | Descriptor: | FORMIC ACID, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
3ZDN
 
 | | D11-C mutant of monoamine oxidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Frank, A, Ghislieri, D, Willies, S, Turner, N.J, Grogan, G. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering an Enantioselective Amine Oxidase for the Synthesis of Pharmaceutical Building Blocks and Alkaloid Natural Products.
J.Am.Chem.Soc., 135, 2013
|
|
7ZKH
 
 | | C-Methyltransferase PsmD from Streptomyces griseofuscus with bound cofactor (crystal form 1) | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, TRIETHYLENE GLYCOL, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-13 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
6NCX
 
 | |
4MXE
 
 | | Human ESCO1 (Eco1/Ctf7 ortholog), acetyltransferase domain in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase ESCO1 | | Authors: | Karlberg, T, Wisniewska, M, Thorsell, A.G, Kouznetsova, E, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Svensson, L, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-26 | | Release date: | 2015-04-08 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sister Chromatid Cohesion Establishment Factor ESCO1 Operates by Substrate-Assisted Catalysis.
Structure, 24, 2016
|
|
1JUH
 
 | | Crystal Structure of Quercetin 2,3-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Schroeter, K.H, Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-08-24 | | Release date: | 2002-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus.
Structure, 10, 2002
|
|
1N0U
 
 | | Crystal structure of yeast elongation factor 2 in complex with sordarin | | Descriptor: | Elongation factor 2, [1R-(1.ALPHA.,3A.BETA.,4.BETA.,4A.BETA.,7.BETA.,7A.ALPHA.,8A.BETA.)]8A-[(6-DEOXY-4-O-METHYL-BETA-D-ALTROPYRANOSYLOXY)METHYL]-4-FORMYL-4,4A,5,6,7,7A,8,8A-OCTAHYDRO-7-METHYL-3-(1-METHYLETHYL)-1,4-METHANO-S-INDACENE-3A(1H)-CARBOXYLIC ACID | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat. Struct. Biol., 10, 2003
|
|
2MLT
 
 | |
