5FPE
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 1H-1,2,4-triazol-3-amine (AT485) in an alternate binding site. | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6I9L
 
 | | JmjC domain-containing protein 5 (JMJD5) in complex with Mn and pyridine-2,4-dicarboxylic acid (2,4-PDCA) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, JmjC domain-containing protein 5, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2018-11-23 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.
Sci Rep, 12, 2022
|
|
7M3P
 
 | | Xrcc4-Spc110p(164-207) fusion | | Descriptor: | Xrcc4-Spc110p(164-207) | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0000186 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
3WOA
 
 | |
5H2X
 
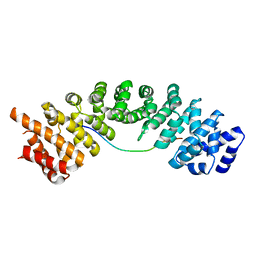 | |
8TEW
 
 | | Human cytomegalovirus penton vertex, CVSC-bound configuration | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
2HBM
 
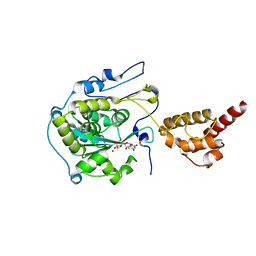 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn, Zn, and UMP | | Descriptor: | Exosome complex exonuclease RRP6, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5FPM
 
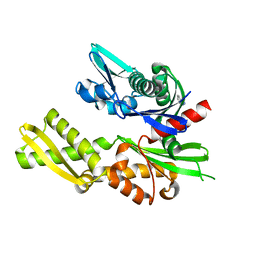 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 5-phenyl-1,3,4-oxadiazole-2-thiol (AT809) in an alternate binding site. | | Descriptor: | 5-PHENYL-1,3,4-OXADIAZOLE-2-THIOL, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KUU
 
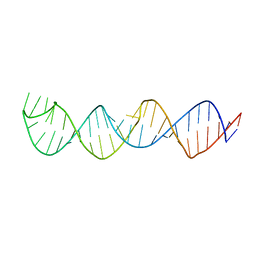 | |
8E2R
 
 | | Crystal structure of TadAC-1.14 | | Descriptor: | GLYCEROL, ZINC ION, tRNA-specific adenosine deaminase 1.14 | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
8E2S
 
 | |
2KW4
 
 | | Solution NMR Structure of the Holo Form of a Ribonuclease H domain from D.hafniense, Northeast Structural Genomics Consortium Target DhR1A | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target DhR1A
To be Published
|
|
2KQ2
 
 | | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A | | Descriptor: | Ribonuclease H-related protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A
To be Published
|
|
8TIA
 
 | |
8TI9
 
 | |
8TI8
 
 | |
6H0U
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with a covalent [1,2,4]triazolo[1,5-a][1,3,5]triazine inhibitor | | Descriptor: | (2~{R})-3-[7-azanyl-5-(cyclohexylamino)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-2-yl]-2-cyano-propanamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marcovich, I, Demitri, N, De Zorzi, R, Storici, P. | | Deposit date: | 2018-07-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Triazolotriazine-Based Dual GSK-3 beta /CK-1 delta Ligand as a Potential Neuroprotective Agent Presenting Two Different Mechanisms of Enzymatic Inhibition.
Chemmedchem, 14, 2019
|
|
5ETB
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED13 | | Descriptor: | 1-(7-methyl-1~{H}-indol-3-yl)ethanone, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
5ETD
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED14 | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
2J4K
 
 | | Crystal structure of uridylate kinase from Sulfolobus solfataricus in complex with UMP to 2.2 Angstrom resolution | | Descriptor: | CADMIUM ION, MAGNESIUM ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Jensen, K.S, Johansson, E, Jensen, K.F. | | Deposit date: | 2006-09-01 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Enzymatic Investigation of the Sulfolobus Solfataricus Uridylate Kinase Shows Competitive Utp Inhibition and the Lack of GTP Stimulation
Biochemistry, 46, 2007
|
|
8U77
 
 | | Crystal structure of Taf14 in complex with Yng1 | | Descriptor: | Protein YNG1, Transcription initiation factor TFIID subunit 14 | | Authors: | Nguyen, M.C, Wei, P.C, Zhang, G.Y, Kutateladze, T.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular insight into interactions between the Taf14, Yng1 and Sas3 subunits of the NuA3 complex.
Nat Commun, 15, 2024
|
|
8U3S
 
 | |
5G4V
 
 | |
8C76
 
 | | Light-state 2.5 Angstrom wild-type X-ray crystal structure of the cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic conditions from a form 2 crystal illuminated during 5 s | | Descriptor: | COBALAMIN, Probable transcriptional regulator | | Authors: | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
5GMK
 
 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | Descriptor: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
