4U5A
 
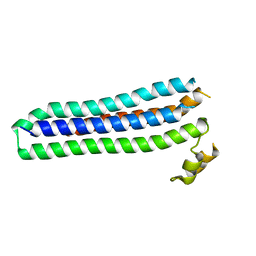 | | Sporozoite Protein for Cell Traversal | | Descriptor: | Sporozoite microneme protein essential for cell traversal | | Authors: | Hamaoka, B.Y. | | Deposit date: | 2014-07-25 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Essential Plasmodium Host Cell Traversal Protein SPECT1.
Plos One, 9, 2014
|
|
1CC2
 
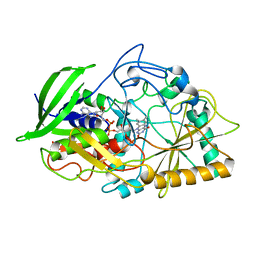 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES HIS447GLN MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
1CDW
 
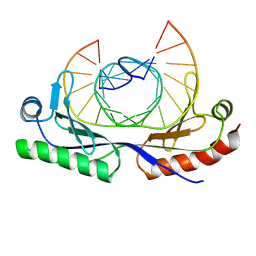 | | HUMAN TBP CORE DOMAIN COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*CP*TP*G)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)) | | Authors: | Nikolov, D.B, Chen, H, Halay, E.D, Hoffmann, A, Roeder, R.G, Burley, S.K. | | Deposit date: | 1996-04-11 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human TATA box-binding protein/TATA element complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4U97
 
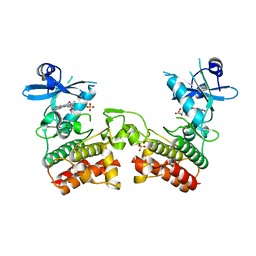 | | Crystal Structure of Asymmetric IRAK4 Dimer | | Descriptor: | Interleukin-1 receptor-associated kinase 4, STAUROSPORINE, SULFATE ION | | Authors: | Ferrao, R, Wu, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | IRAK4 Dimerization and trans-Autophosphorylation Are Induced by Myddosome Assembly.
Mol.Cell, 55, 2014
|
|
4U6I
 
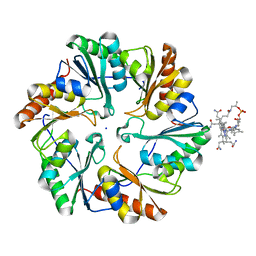 | | Crystal Structure of the EutL Microcompartment Shell Protein from Clostridium Perfringens Bound to Vitamin B12 | | Descriptor: | COBALAMIN, Ethanolamine utilization protein EutL, SODIUM ION | | Authors: | Thompson, M.C, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1C8X
 
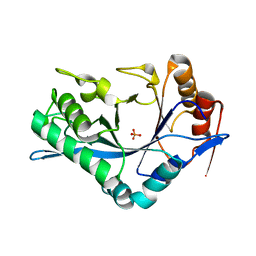 | | Endo-Beta-N-Acetylglucosaminidase H, D130E Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, PHOSPHATE ION | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1CA4
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 (TRAF2) | | Descriptor: | PROTEIN (TNF RECEPTOR ASSOCIATED FACTOR 2) | | Authors: | Park, Y.C, Burkitt, V, Villa, A.R, Tong, L, Wu, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for self-association and receptor recognition of human TRAF2.
Nature, 398, 1999
|
|
1C91
 
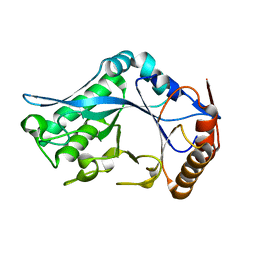 | | Endo-Beta-N-Acetylglucosaminidase H, E132D | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C9C
 
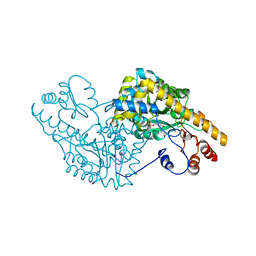 | | ASPARTATE AMINOTRANSFERASE COMPLEXED WITH C3-PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1CCP
 
 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
3N2J
 
 | | Azurin H117G, oxidized form | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Hoffmann, M, Alagaratnam, S, Canters, G.W, Einsle, O. | | Deposit date: | 2010-05-18 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Probing the reactivity of different forms of azurin by flavin photoreduction.
Febs J., 278, 2011
|
|
4U9A
 
 | |
1C8Y
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ZINC ION | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1CCZ
 
 | | CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CD58) | | Authors: | Ikemizu, S, Sparks, L.M, Van Der Merwe, P.A, Harlos, K, Stuart, D.I, Jones, E.Y, Davis, S.J. | | Deposit date: | 1999-03-02 | | Release date: | 1999-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C7K
 
 | | CRYSTAL STRUCTURE OF THE ZINC PROTEASE | | Descriptor: | CALCIUM ION, ZINC ENDOPROTEASE, ZINC ION | | Authors: | Kurisu, G, Harada, S, Kai, Y. | | Deposit date: | 2000-02-19 | | Release date: | 2001-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 A resolution.
J.Inorg.Biochem., 82, 2000
|
|
1CWH
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 3-D-SER CYCLOSPORIN | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-07 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1C9D
 
 | |
5E4J
 
 | | Acetylcholinesterase Methylene Blue no PEG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, DECAMETHONIUM ION, ... | | Authors: | Dym, O. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-30 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The impact of crystallization conditions on structure-based drug design: A case study on the methylene blue/acetylcholinesterase complex.
Protein Sci., 25, 2016
|
|
1C8V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-30 | | Release date: | 2000-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1C7R
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | 5-PHOSPHOARABINONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
1C9M
 
 | | BACILLUS LENTUS SUBTILSIN (SER 87) N76D/S103A/V104I | | Descriptor: | CALCIUM ION, SERINE PROTEASE, SULFATE ION | | Authors: | Bott, R. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Engineered Bacillus lentus subtilisins having altered flexibility.
J.Mol.Biol., 292, 1999
|
|
1C7Q
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | N-BROMOACETYL-AMINOETHYL PHOSPHATE, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
4U7M
 
 | | LRIG1 extracellular domain: Structure and Function Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeats and immunoglobulin-like domains protein 1 | | Authors: | Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | LRIG1 Extracellular Domain: Structure and Function Analysis.
J.Mol.Biol., 427, 2015
|
|
4UFY
 
 | | Crystal structure of human tankyrase 2 in complex with TA-13 | | Descriptor: | 8-METHYL-2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UE2
 
 | | Structure of air-treated anaerobically purified D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-15 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | [Nife]-Hydrogenases Revisited: Nickel-Carboxamido Bond Formation in a Variant with Accrued O2-Tolerance and a Tentative Re-Interpretation of Ni-Si States.
Metallomics, 7, 2015
|
|
