7KOB
 
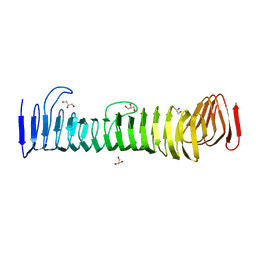 | | Crystal structure of antigen 43b from Escherichia coli CFT073 | | Descriptor: | Antigen 43b, GLYCEROL | | Authors: | Martinez-Ortiz, G.C, Hor, L, Paxman, J.J, Heras, B. | | Deposit date: | 2020-11-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms.
NPJ Biofilms Microbiomes, 8, 2022
|
|
3H1P
 
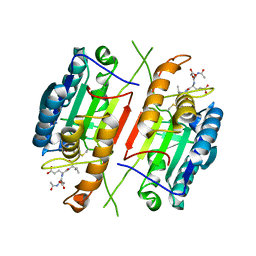 | |
7KOH
 
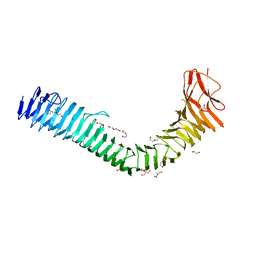 | | Crystal structure of antigen 43 from Escherichia coli EDL933 | | Descriptor: | Antigen 43, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vo, J.L, Paxman, J.J, Heras, B. | | Deposit date: | 2020-11-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms.
NPJ Biofilms Microbiomes, 8, 2022
|
|
7ICM
 
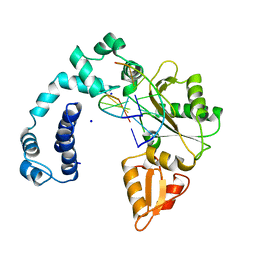 | |
7JJJ
 
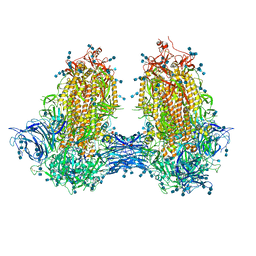 | | Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7FAW
 
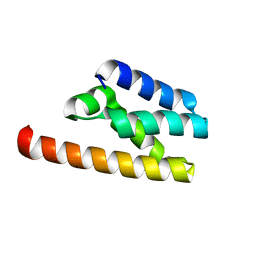 | | Structure of LW domain from Yeast | | Descriptor: | Transcription elongation factor S-II | | Authors: | Liao, S, Gao, J, Tu, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.
Int.J.Biol.Macromol., 253, 2023
|
|
3GQ0
 
 | |
3AX0
 
 | |
7FBL
 
 | | Thrombocorticin in complex with Ca2+ and mannose | | Descriptor: | CALCIUM ION, Thrombocorticin, alpha-D-mannopyranose | | Authors: | Kageyama, H, Onodera, K, Sakai, R, Tanaka, Y. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | A marine sponge-derived lectin reveals hidden pathway for thrombopoietin receptor activation.
Nat Commun, 13, 2022
|
|
3AWZ
 
 | |
2P2O
 
 | | Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Maltose Transacetylase from Geobacillus Kaustophilus at 1.8 Angstrom Resolution
To be Published
|
|
3AWU
 
 | |
7FBI
 
 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
3AWX
 
 | |
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
2JLC
 
 | | Crystal structure of E.coli MenD, 2-succinyl-5-enolpyruvyl-6-hydroxy- 3-cyclohexadiene-1-carboxylate synthase - native protein | | Descriptor: | 2-SUCCINYL-5-ENOLPYRUVYL-6-HYDROXY-3-CYCLOHEXENE -1-CARBOXYLATE SYNTHASE, MANGANESE (II) ION, THIAMINE DIPHOSPHATE | | Authors: | Dawson, A, Fyfe, P.K, Hunter, W.N. | | Deposit date: | 2008-09-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity and Reactivity in Menaquinone Biosynthesis: The Structure of Escherichia Coli Mend (2-Succinyl-5-Enolpyruvyl-6-Hydroxy-3-Cyclohexadiene-1-Carboxylate Synthase).
J.Mol.Biol., 384, 2008
|
|
2PVL
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-ETHYLPIPERAZIN-1-YL)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JD7
 
 | |
2JJ6
 
 | | Crystal structure of the putative carbohydrate recognition domain of the human galectin-related protein | | Descriptor: | GALECTIN-RELATED PROTEIN | | Authors: | Thore, S, Walti, M.A, Kunzler, M, Aebi, M. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Carbohydrate Recognition Domain of Human Galectin-Related Protein
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
3GVD
 
 | | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETIC ACID, CYSTEINE, ... | | Authors: | Kim, Y, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-30 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis
To be Published
|
|
2PWC
 
 | | HIV-1 protease in complex with a amino decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, Gag-Pol polyprotein (Pr160Gag-Pol), ... | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-11 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
3H8X
 
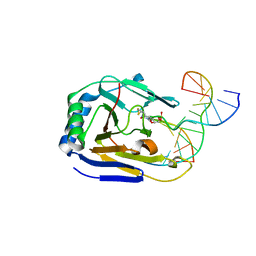 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | Descriptor: | 5'-D(*CP*TP*GP*TP*(ME6)P*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*AP*TP*AP*AP*GP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 | | Authors: | Lu, L, Yi, C, Jian, X, Zheng, G, He, C. | | Deposit date: | 2009-04-29 | | Release date: | 2010-03-31 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
3AXM
 
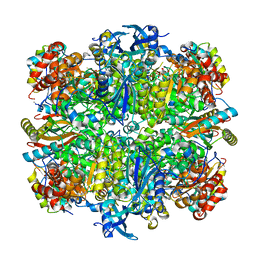 | | Structure of rice Rubisco in complex with 6PG | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
2JHJ
 
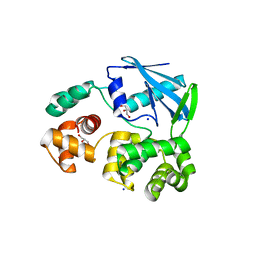 | | 3-methyladenine dna-glycosylase from Archaeoglobus fulgidus | | Descriptor: | 3-METHYLADENINE DNA-GLYCOSYLASE, GLYCEROL, SODIUM ION | | Authors: | Leiros, I, Nabong, M.P, Grosvik, K, Ringvoll, J, Haugland, G.T, Uldal, L, Reite, K, Olsbu, I.K, Knaevelsrud, I, Moe, E, Andersen, O.A, Birkeland, N.K, Ruoff, P, Klungland, A, Bjelland, S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Enzymatic Excision of N1-Methyladenine and N3-Methylcytosine from DNA
Embo J., 26, 2007
|
|
7FCY
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | (1S)-2-methyl-1-(2-methyl-1,2,4-triazol-3-yl)propan-1-amine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
