2CZU
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
2CZV
 
 | | Crystal structure of archeal RNase P protein ph1481p in complex with ph1877p | | Descriptor: | ACETIC ACID, Ribonuclease P protein component 2, Ribonuclease P protein component 3, ... | | Authors: | Kawano, S, Kakuta, Y, Nakashima, T, Tanaka, I, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme
J.Mol.Biol., 357, 2006
|
|
2CZW
 
 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
2D00
 
 | | Subunit F of V-type ATPase/synthase | | Descriptor: | CALCIUM ION, V-type ATP synthase subunit F | | Authors: | Makyio, H, Iino, R, Ikeda, C, Imamura, H, Tamakoshi, M, Iwata, M, Stock, D, Bernal, R.A, Carpenter, E.P, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2005-07-21 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus
Embo J., 24, 2005
|
|
2D01
 
 | | Wild Type Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
2D02
 
 | | R52Q Mutant of Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
2D03
 
 | | Crystal structure of the G91S mutant of the NNA7 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Xie, K, Song, S.C, Spitalnik, S.L, Wedekind, J.E. | | Deposit date: | 2005-07-23 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic analysis of the NNA7 Fab and proposal for the mode of human blood-group recognition.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2D04
 
 | | Crystal structure of neoculin, a sweet protein with taste-modifying activity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Curculin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimizu-Ibuka, A, Morita, Y, Terada, T, Asakura, T, Nakajima, K, Iwata, S, Misaka, T, Sorimachi, H, Arai, S, Abe, K. | | Deposit date: | 2005-07-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of neoculin: insights into its sweetness and taste-modifying activity
J.Mol.Biol., 359, 2006
|
|
2D05
 
 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
2D06
 
 | | Human Sult1A1 Complexed With Pap and estradiol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ESTRADIOL, Sulfotransferase 1A1 | | Authors: | Gamage, N.U, Tsvetanov, S, Duggleby, R.G, McManus, M.E, Martin, J.L. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi-ring substrates
J.Biol.Chem., 280, 2005
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
2D09
 
 | |
2D0A
 
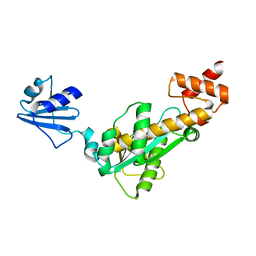 | | Crystal structure of Bst-RNase HIII | | Descriptor: | ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0B
 
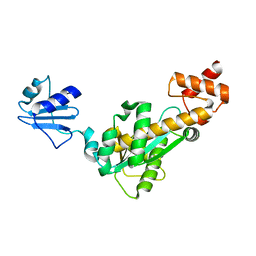 | | Crystal structure of Bst-RNase HIII in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0C
 
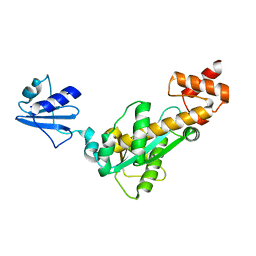 | | Crystal structure of Bst-RNase HIII in complex with Mn2+ | | Descriptor: | MANGANESE (II) ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0D
 
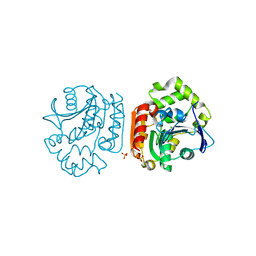 | | Crystal Structure of a Meta-cleavage Product Hydrolase (CumD) A129V Mutant | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, CHLORIDE ION, PHOSPHATE ION | | Authors: | Jun, S.Y, Fushinobu, S, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2005-08-01 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improving the catalytic efficiency of a meta-cleavage product hydrolase (CumD) from Pseudomonas fluorescens IP01
Biochim.Biophys.Acta, 1764, 2006
|
|
2D0E
 
 | | Substrate assited in Oxygen Activation in Cytochrome P450 158A2 | | Descriptor: | 2-HYDROXYNAPHTHOQUINONE, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2005-08-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of active site water molecules and substrate hydroxyl groups in oxygen activation by cytochrome P450 158A2: a new mechanism of proton transfer
J.Biol.Chem., 280, 2005
|
|
2D0F
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D0G
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D0H
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D0I
 
 | |
2D0J
 
 | | Crystal Structure of Human GlcAT-S Apo Form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 | | Authors: | Shiba, T, Kakuda, S, Ishiguro, M, Oka, S, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-08-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GlcAT-S, a human glucuronyltransferase, involved in the biosynthesis of the HNK-1 carbohydrate epitope
To be published
|
|
2D0K
 
 | |
2D0N
 
 | |
