2QVR
 
 | | E. coli Fructose-1,6-bisphosphatase: Citrate, Fru-2,6-P2, and Mg2+ bound | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CITRIC ACID, Fructose-1,6-bisphosphatase, ... | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of Mammalian and Bacterial Fructose-1,6-bisphosphatase Reveal the Basis for Synergism in AMP/Fructose 2,6-Bisphosphate Inhibition.
J.Biol.Chem., 282, 2007
|
|
5ERS
 
 | | GephE in complex with Mg(2+) - AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2015-11-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Framework for Metal Incorporation during Molybdenum Cofactor Biosynthesis.
Structure, 24, 2016
|
|
6ANG
 
 | |
4CVM
 
 | |
4KM5
 
 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
1MF0
 
 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP, GDP, HPO4(2-), and Mg(2+) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1MF1
 
 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
2GXQ
 
 | | HERA N-terminal domain in complex with AMP, crystal form 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, heat resistant RNA dependent ATPase | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2006-05-09 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure and Nucleotide Binding of the Thermus thermophilus RNA Helicase Hera N-terminal Domain.
J.Mol.Biol., 351, 2006
|
|
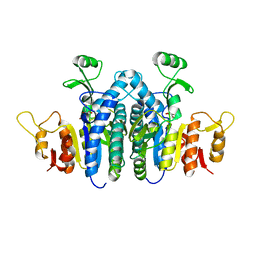
4JZ9
 
 | |
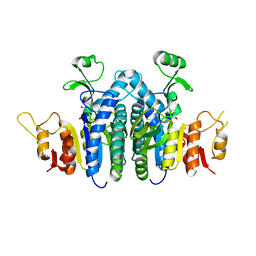
4JZ8
 
 | |
1FJ9
 
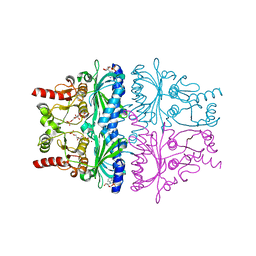 | | FRUCTOSE-1,6-BISPHOSPHATASE (MUTANT Y57W) PRODUCTS/ZN/AMP COMPLEX (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Iancu, C.V, Choe, J.Y, Honzatko, R.B. | | Deposit date: | 2000-08-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tryptophan fluorescence reveals the conformational state of a dynamic loop in recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry, 39, 2000
|
|
5U7V
 
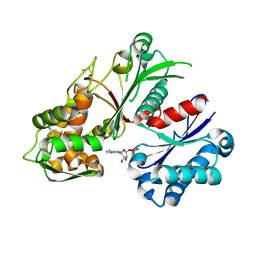 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Apyrase | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
8XBK
 
 | | Crystal Structure of Human Liver Fructose-1,6-bisphosphatase Complexed with a Covalent Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Cao, H, Zhang, X, Ren, Y, Wan, J. | | Deposit date: | 2023-12-06 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-Guided Design of Affinity/Covalent-Bond Dual-Driven Inhibitors Targeting the AMP Site of FBPase.
J.Med.Chem., 67, 2024
|
|
5VN4
 
 | |
7CTV
 
 | |
7CTX
 
 | |
4HE1
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with fructose-6-phosphate and phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3L2B
 
 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
2QRK
 
 | | Crystal Structure of AMP-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Saccharopine dehydrogenase [NAD+, L-lysine-forming | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
6EA9
 
 | |
7VGF
 
 | | cryo-EM structure of AMP-PNP bound human ABCB7 | | Descriptor: | Iron-sulfur clusters transporter ABCB7, mitochondrial, MAGNESIUM ION, ... | | Authors: | Yan, Q, Yang, X, Shen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-02-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of AMP-PNP-bound human mitochondrial ATP-binding cassette transporter ABCB7.
J.Struct.Biol., 214, 2022
|
|
7ELT
 
 | |
6Z0Z
 
 | | Human wtSTING in complex with 3',3'-c-(2'FdAMP-2'FdAMP) | | Descriptor: | 2'-fluoro-,3',3'-c-di-AMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1Z6S
 
 | | Ribonuclease A- AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Hatzopoulos, G.N, Leonidas, D.D, Kardakaris, R, Kobe, J, Oikonomakos, N.G. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The binding of IMP to Ribonuclease A
Febs J., 272, 2005
|
|
3BA6
 
 | | Structure of the Ca2E1P phosphoenzyme intermediate of the SERCA Ca2+-ATPase | | Descriptor: | AMP PHOSPHORAMIDATE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Picard, M, Winther, A.M.L, Olesen, C, Gyrup, C, Morth, J.P, Oxvig, C, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of calcium transport by the calcium pump.
Nature, 450, 2007
|
|
