5OTQ
 
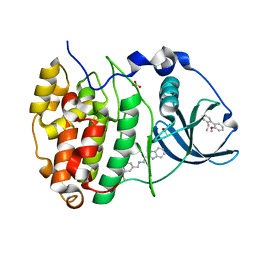 | | The crystal structure of CK2alpha in complex with compound 33 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[4-(2-ethylphenyl)-3-methoxy-phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
5OTZ
 
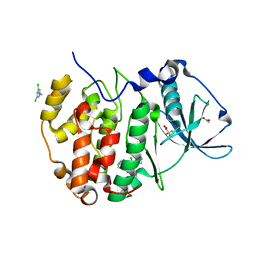 | | The crystal structure of CK2alpha in complex with compound 1 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, [3,5-bis(chloranyl)-4-(2-ethylphenyl)phenyl]methanamine, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1WMM
 
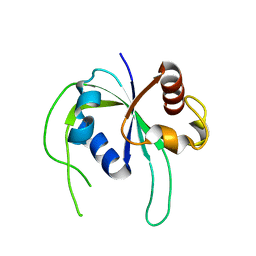 | |
4BNX
 
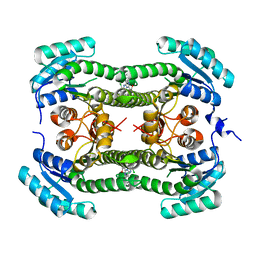 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 6-(4-(2-chloroanilino)- 1H-quinazolin-2-ylidene)cyclohexa-2, 4-dien-1-one at 2.3A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 6-[4-(2-chloroanilino)-1H-quinazolin-2-ylidene]cyclohexa-2,4-dien-1-one | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO3
 
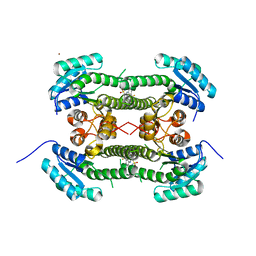 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-(3-(trifluoromethyl) anilino)pyridine-3-sulfonamide at 2.5A resolution | | Descriptor: | 2-(3-(trifluoromethyl)anilino)pyridine-3-sulfonamide, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
3VGD
 
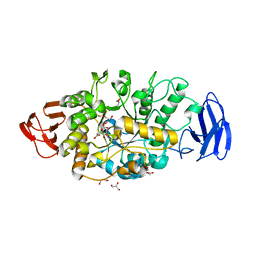 | | Ctystal structure of glycosyltrehalose trehalohydrolase (D252E) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
4G3J
 
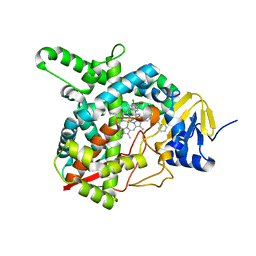 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the VNI derivative (R)-N-(1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide [R-VNI-triazole (VNT)] | | Descriptor: | N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2012-07-14 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | VFV as a New Effective CYP51 Structure-Derived Drug Candidate for Chagas Disease and Visceral Leishmaniasis.
J Infect Dis, 212, 2015
|
|
4BS0
 
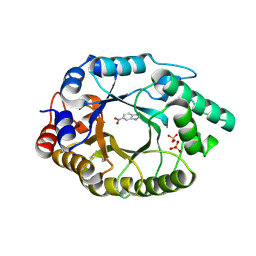 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
2BS2
 
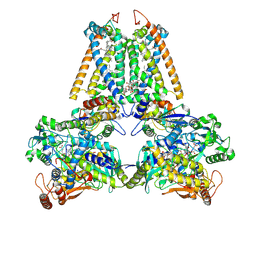 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Evidence for Transmembrane Proton Transfer in a Dihaem-Containing Membrane Protein Complex.
Embo J., 25, 2006
|
|
4BTH
 
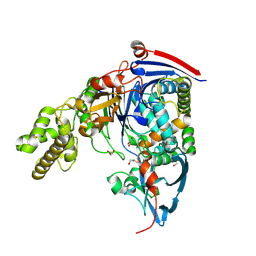 | | The LeuA146Trp,PheB24Tyr Double Mutant of the Quorum Quenching N-acyl Homoserine Lactone Acylase PvdQ Has an Altered Substrate Specificity Towards Small Acyl Chains | | Descriptor: | ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, GLYCEROL | | Authors: | Koch, G, Nadal-Jimenez, P, Reis, C.R, Muntendam, R, Bokhove, M, Melillo, E, Dijkstra, B.W, Cool, R.H, Quax, W.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reducing Virulence of the Human Pathogen Burkholderia by Altering the Substrate Specificity of the Quorum-Quenching Acylase Pvdq
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2C3O
 
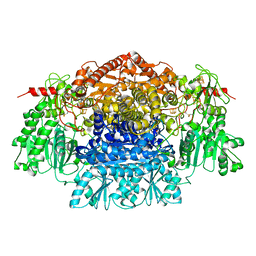 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3U
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus, Oxygen inhibited form | | Descriptor: | 2-(3-{[4-(HYDROXYAMINO)-2-METHYLPYRIMIDIN-5-YL]METHYL}-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL)ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C42
 
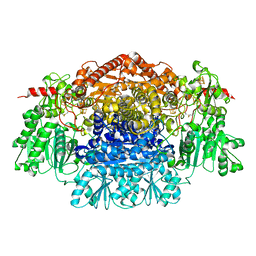 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-14 | | Release date: | 2006-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
4BQ3
 
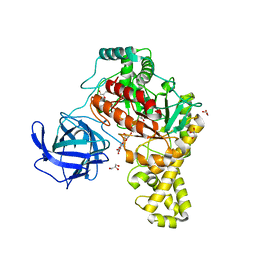 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
3VIM
 
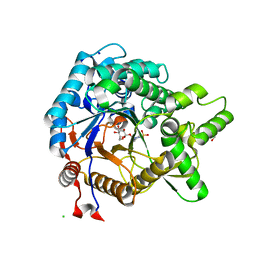 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product | | Descriptor: | 2-{4-[2-(beta-D-glucopyranosyloxy)ethyl]piperazin-1-yl}ethanesulfonic acid, Beta-glucosidase, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1NQO
 
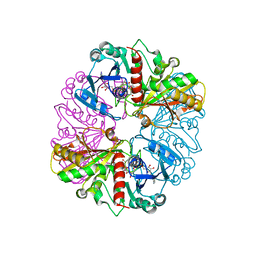 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate
Dehydrogenase from Bacillus stearothermophilus with NAD and
D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
4G7G
 
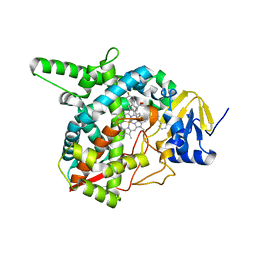 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the VNI derivative (R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide [VNI/VNF (VFV)] | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2012-07-20 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | VFV as a New Effective CYP51 Structure-Derived Drug Candidate for Chagas Disease and Visceral Leishmaniasis.
J Infect Dis, 212, 2015
|
|
5OWI
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 1) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
1O0S
 
 | | Crystal Structure of Ascaris suum Malic Enzyme Complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD-dependent malic enzyme, TARTRONATE | | Authors: | Rao, G.S, Coleman, D.E, Karsten, W.E, Cook, P.F, Harris, B.G. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site.
J.Biol.Chem., 278, 2003
|
|
1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF, SULFATE ION | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-30 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
3VM5
 
 | |
2Y6L
 
 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
3VML
 
 | |
4BO9
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 5-(2-(furan-2-ylmethoxy) phenyl)-2-phenyltetrazole at 2.9A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 5-[2-(FURAN-2-YLMETHOXY)PHENYL]-2-PHENYLTETRAZOLE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
2C41
 
 | | X-ray structure of Dps from Thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, DPS FAMILY DNA-BINDING STRESS RESPONSE PROTEIN, TETRAETHYLENE GLYCOL, ... | | Authors: | Ilari, A, Franceschini, S, Ceci, P, Chiancone, E. | | Deposit date: | 2005-10-14 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Antioxidant Dps Protein from the Thermophilic Cyanobacterium Thermosynechococcus Elongatus.
FEBS J., 273, 2006
|
|
