7BGL
 
 | | Salmonella LP ring 26 mer refined in C26 map | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Flagellar L-ring protein, Flagellar P-ring protein, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-07 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
1E28
 
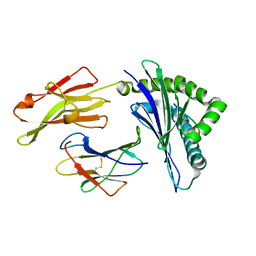 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM2(TAFTIPSI) | | Descriptor: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN, PEPTIDE | | Authors: | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
7BIN
 
 | | Salmonella export gate and rod refined in focussed C1 map | | Descriptor: | Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, Flagellar basal-body rod protein FlgF, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-12 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
5LTQ
 
 | |
1E2W
 
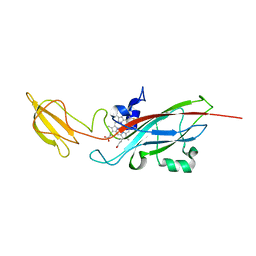 | | N168F mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
7BL4
 
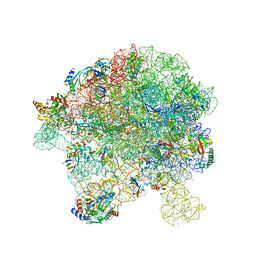 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL6
 
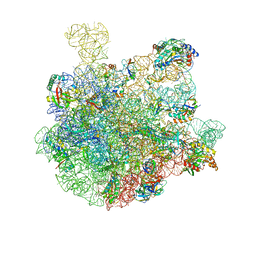 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
5FGA
 
 | |
7BL5
 
 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
1DMI
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 6S-H4B | | Descriptor: | 6S-5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kotsonis, P, Pfleiderer, W, Schmidt, H.H.H.W, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bovine Endothelial Nitric Oxide Synthase Heme Domain Complexed with the Pterin Analogues
To be Published
|
|
1DWH
 
 | |
1DO3
 
 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) AFTER PHOTOLYSIS AT T>180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1DWW
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DOC
 
 | | THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, ... | | Authors: | Gatti, D.L, Palfey, B.A, Lah, M.S, Entsch, B, Massey, V, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mobile flavin of 4-OH benzoate hydroxylase.
Science, 266, 1994
|
|
1DZJ
 
 | | Porcine Odorant Binding Protein Complexed with 2-amino-4-butyl-5-propylselenazole | | Descriptor: | 4-butyl-5-propyl-1,3-selenazol-2-amine, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-01 | | Release date: | 2000-12-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1DOK
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1DZQ
 
 | | LECTIN UEA-II COMPLEXED WITH GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN II, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 2000-03-07 | | Release date: | 2000-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
7BGN
 
 | |
1DQH
 
 | |
5LZT
 
 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
7B93
 
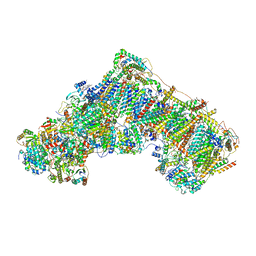 | | Cryo-EM structure of mitochondrial complex I from Mus musculus inhibited by IACS-2858 at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1-[[3-(4-methylsulfonylpiperidin-1-yl)phenyl]methyl]-5-[3-[4-(trifluoromethyloxy)phenyl]-1,2,4-oxadiazol-5-yl]pyridin-2-one, ... | | Authors: | Chung, I, Hirst, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cork-in-bottle mechanism of inhibitor binding to mammalian complex I.
Sci Adv, 7, 2021
|
|
1DTH
 
 | | METALLOPROTEASE | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ATROLYSIN C, CALCIUM ION, ... | | Authors: | Botos, I, Scapozza, L, Zhang, D, Liotta, L.A, Meyer, E.F. | | Deposit date: | 1996-02-12 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Batimastat, a potent matrix mealloproteinase inhibitor, exhibits an unexpected mode of binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1DKN
 
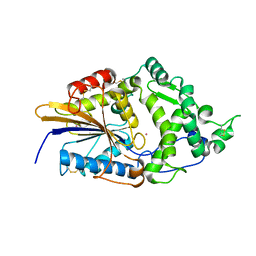 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 5.0 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DVJ
 
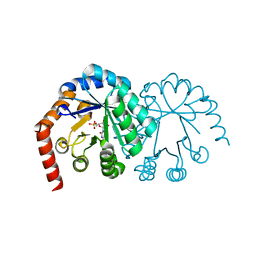 | | CRYSTAL STRUCTURE OF OROTIDINE MONOPHOSPHATE DECARBOXYLASE COMPLEXED WITH 6-AZAUMP | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Wu, N, Mo, Y, Gao, J, Pai, E.F. | | Deposit date: | 2000-03-30 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DWQ
 
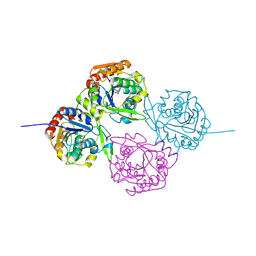 | | Crystal Structure of Hydroxynitrile Lyase from Manihot esculenta in Complex with Substrates Acetone and Chloroacetone:Implications for the Mechanism of Cyanogenesis | | Descriptor: | CHLOROACETONE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Forster, S, Mielich, B, Wajant, H, Effenberger, F. | | Deposit date: | 1999-12-10 | | Release date: | 2000-12-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Substrates Acetone and Chloroacetone: Implications for the Mechanism of Cyanogenesis
Acta Crystallogr.,Sect.D, 57, 2001
|
|
