5LAT
 
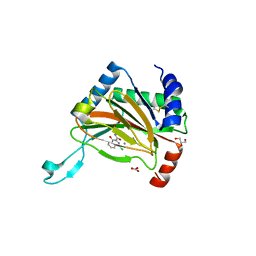 | |
3ISE
 
 | | Structure of mineralized Bfrb (double soak) from Pseudomonas aeruginosa to 2.8A Resolution | | Descriptor: | Bacterioferritin, FE (III) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
7TAK
 
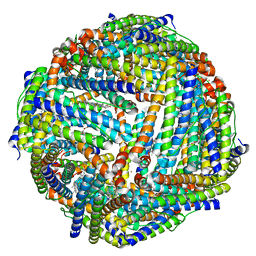
 | | Structure of a NAT transporter | | Descriptor: | GUANINE, Putative membrane protein PurT | | Authors: | Weng, J, Zhou, X, Ren, Z, Chen, K, Zhou, M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.79828 Å) | | Cite: | Insight into the mechanism of H + -coupled nucleobase transport.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PO5
 
 | | Lactobacillus plantarum LpdD | | Descriptor: | MANGANESE (II) ION, Protein LpdD | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gallate decarboxylase subunit D, LpdD
To Be Published
|
|
7QHM
 
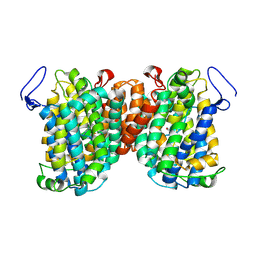
 | | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound) | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, AZIDE ION, ... | | Authors: | Kao, W.-C, Hunte, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for safe and efficient energy conversion in a respiratory supercomplex.
Nat Commun, 13, 2022
|
|
8PZH
 
 | | LpdD (H61A) mutant | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein LpdD | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of LpdD (H61A) mutant from Lactobacillus plantarum.
To Be Published
|
|
7QHO
 
 | | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated) | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Kao, W.-C, Hunte, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for safe and efficient energy conversion in a respiratory supercomplex
Nature Communications, 13, 2022
|
|
7TEK
 
 | |
2IL8
 
 | |
3IOF
 
 | | Crystal structure of CphA N220G mutant with inhibitor 10a | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION, ... | | Authors: | Delbruck, H, Bebrone, C, Hoffmann, K.M.V. | | Deposit date: | 2009-08-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-beta-lactamases
J.Med.Chem., 53, 2010
|
|
3IUD
 
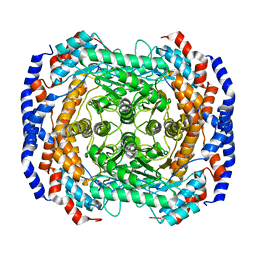 | | Cu2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COPPER (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
8Q62
 
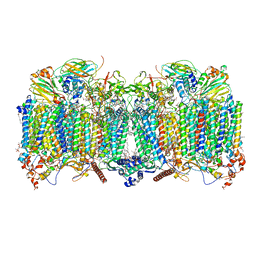
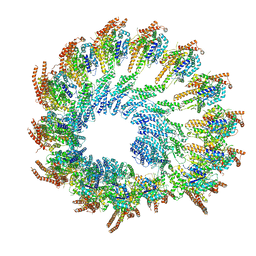 | | Early closed conformation of the g-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 2, Gamma-tubulin complex component 3, Gamma-tubulin complex component 4, ... | | Authors: | Llorca, O, Serna, M, Fernandez-Leiro, R. | | Deposit date: | 2023-08-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Transition of human gamma-tubulin ring complex into a closed conformation during microtubule nucleation.
Science, 383, 2024
|
|
7FEM
 
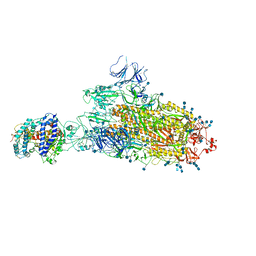 | | SARS-CoV-2 B.1.1.7 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wen, Z.L, Zhu, Y, Sun, F. | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure-based evidence for the enhanced transmissibility of the dominant SARS-CoV-2 B.1.1.7 variant (Alpha).
Cell Discov, 7, 2021
|
|
7T23
 
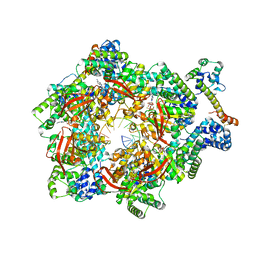 | | E. coli DnaB bound to two DnaG C-terminal domains, ssDNA, ADP and AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase, ... | | Authors: | Oakley, A.J, Xu, Z.Q. | | Deposit date: | 2021-12-02 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sequence of conformation changes of DnaB helicase during DNA unwinding and priming in Escherichia coli
To Be Published
|
|
8PH4
 
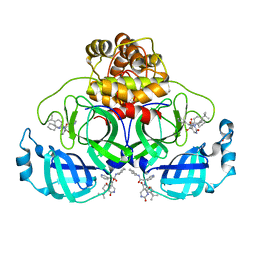 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
7TEL
 
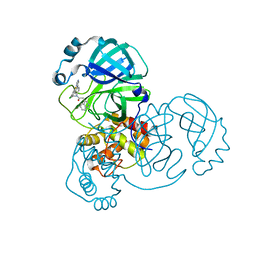 | |
5L6M
 
 | | Structure of Caulobacter crescentus VapBC1 (VapB1deltaC:VapC1 form) | | Descriptor: | GLYCEROL, MALONATE ION, Ribonuclease VapC, ... | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
2XTM
 
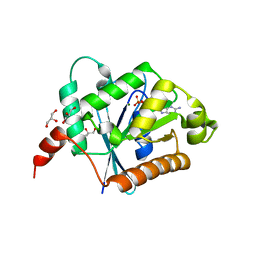 | | Crystal structure of GDP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GLYCEROL, GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7FHR
 
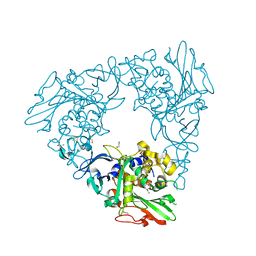 | | Crystal Structure of a Rieske Oxygenase from Cupriavidus metallidurans | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mahto, J.K, Dhankhar, P, Kumar, P. | | Deposit date: | 2021-07-30 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by phthalate dioxygenase from Comamonas testosteroni.
J.Biol.Chem., 297, 2021
|
|
7TC7
 
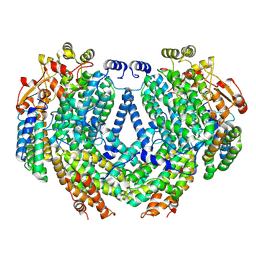 | | Cryo-EM structure of methane monooxygenase hydroxylase (by quantifoil) | | Descriptor: | FE (III) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Cho, U.S, Kim, B.C. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Batch Production of High-Quality Graphene Grids for Cryo-EM: Cryo-EM Structure of Methylococcus capsulatus Soluble Methane Monooxygenase Hydroxylase.
Acs Nano, 17, 2023
|
|
5L6V
 
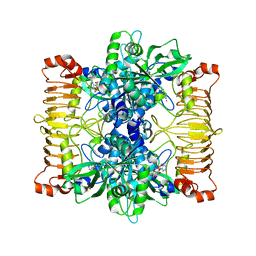 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.667 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
3IVY
 
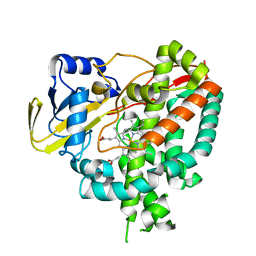 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125, p212121 crystal form | | Descriptor: | Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
2XRP
 
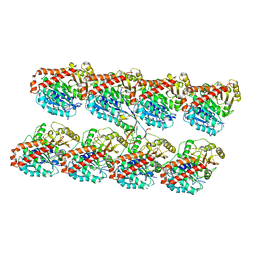 | | Human Doublecortin N-DC Repeat (1MJD) and Mammalian Tubulin (1JFF and 3HKE) Docked into the 8-Angstrom Cryo-EM Map of Doublecortin- Stabilised Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, NEURONAL MIGRATION PROTEIN DOUBLECORTIN, ... | | Authors: | Fourniol, F.J, Sindelar, C.V, Amigues, B, Clare, D.K, Thomas, G, Perderiset, M, Francis, F, Houdusse, A, Moores, C.A. | | Deposit date: | 2010-09-18 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Template-Free 13-Protofilament Microtubule-Map Assembly Visualized at 8 A Resolution.
J.Cell Biol., 191, 2010
|
|
7AKT
 
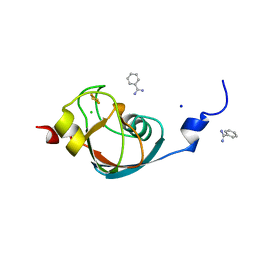 | | CrPetF variant - A39G_A41V | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kurisu, G, Ohnishi, Y, Engelbrecht, V, Happe, T. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ferredoxin 2.0: an electron transfer protein designed into a photosystem I-driven hydrogenase
To Be Published
|
|
2XSN
 
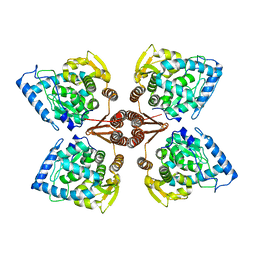 | | Crystal Structure of Human Tyrosine Hydroxylase Catalytic Domain | | Descriptor: | TYROSINE 3-MONOOXYGENASE, ZINC ION | | Authors: | Muniz, J.R.C, Cooper, C.D.O, Yue, W.W, Krysztofinska, E, von Delft, F, Knapp, S, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-10-29 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of Human Tyrosine Hydroxylase Catalytic Domain
To be Published
|
|
