1NXY
 
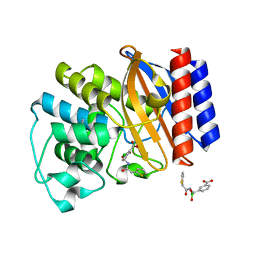 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (SM2) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase TEM, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
5QK0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1270312110 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,5-dimethyl-1H-pyrazol-4-yl)aniline, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2UUE
 
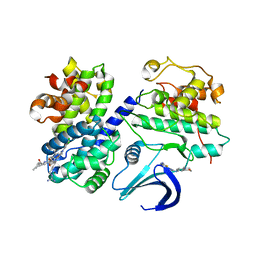 | | REPLACE: A strategy for Iterative Design of Cyclin Binding Groove Inhibitors | | Descriptor: | 1-(3,5-DICHLOROPHENYL)-5-METHYL-1H-1,2,4-TRIAZOLE-3-CARBOXYLIC ACID, 4-METHYL-5-{(2E)-2-[(4-MORPHOLIN-4-YLPHENYL)IMINO]-2,5-DIHYDROPYRIMIDIN-4-YL}-1,3-THIAZOL-2-AMINE, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Andrews, M.J, Kontopidis, G, McInnes, C, Plater, A, Innes, L, Cowan, A, Jewsbury, P, Fischer, P.M. | | Deposit date: | 2007-03-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Replace: A Strategy for Iterative Design of Cyclin- Binding Groove Inhibitors
Chembiochem, 7, 2006
|
|
3T1L
 
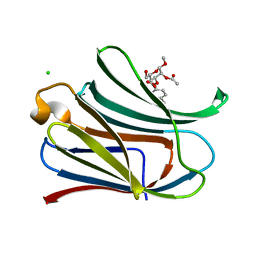 | |
5QTM
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with FS-2639 | | Descriptor: | 1,2-ETHANEDIOL, 7-fluoroquinazolin-4(3H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2X7G
 
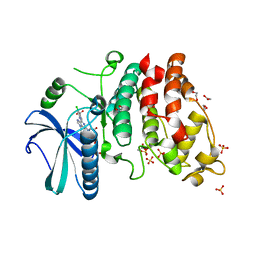 | | Structure of human serine-arginine-rich protein-specific kinase 2 (SRPK2) bound to purvalanol B | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PURVALANOL B, ... | | Authors: | Pike, A.C.W, Savitsky, P, Fedorov, O, Krojer, T, Ugochukwu, E, von Delft, F, Gileadi, O, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Serine-Arginine-Rich Protein- Specific Kinase 2 (Srpk2) Bound to Purvalanol B
To be Published
|
|
5QOO
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000144a | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-{5-[(2S)-oxolan-2-yl]-1,3,4-thiadiazol-2(3H)-ylidene}propanamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4E77
 
 | | 2.0A Crystal Structure of a Glutamate-1-Semialdehyde Aminotransferase from Yersinia pestis CO92 | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase, NITRATE ION, SODIUM ION | | Authors: | Brunzelle, J.S, Wawrzak, W, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-16 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A Crystal Structure of a Glutamate-1-Semialdehyde Aminotransferase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
4EL6
 
 | | Crystal structure of IPSE/alpha-1 from Schistosoma mansoni eggs | | Descriptor: | IL-4-inducing protein | | Authors: | Mayerhofer, H, Meyer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Sattler, M, Schramm, G, Mueller-Dieckmann, J. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A Crystallin Fold in the Interleukin-4-inducing Principle of Schistosoma mansoni Eggs (IPSE/ alpha-1) Mediates IgE Binding for Antigen-independent Basophil Activation.
J.Biol.Chem., 290, 2015
|
|
5QPA
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000449a | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1FLP
 
 | | STRUCTURE OF THE SULFIDE-REACTIVE HEMOGLOBIN FROM THE CLAM LUCINA PECTINATA: CRYSTALLOGRAPHIC ANALYSIS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN I (AQUO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rizzi, M, Wittenberg, J.B, Ascenzi, P, Fasano, M, Coda, A, Bolognesi, M. | | Deposit date: | 1994-05-16 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sulfide-reactive hemoglobin from the clam Lucina pectinata. Crystallographic analysis at 1.5 A resolution.
J.Mol.Biol., 244, 1994
|
|
5QTS
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with 8J-537S | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-(methylsulfanyl)-6-(trifluoromethyl)pyrimidin-4(3H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
193L
 
 | | THE 1.33 A STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4ITX
 
 | | P113S mutant of E. coli Cystathionine beta-lyase MetC inhibited by reaction with L-Ala-P | | Descriptor: | CALCIUM ION, Cystathionine beta-lyase MetC, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Squire, C.J, Yosaatmadja, Y, Soo, V.W.C, Patrick, W.M. | | Deposit date: | 2013-01-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
7F4D
 
 | | Cryo-EM structure of alpha-MSH-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4I
 
 | | Cryo-EM structure of SHU9119-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
1SFT
 
 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, Alanine racemase | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
4IYE
 
 | | Crystal structure of AdTx1 (rho-Da1a) from eastern green mamba (Dendroaspis angusticeps) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Toxin AdTx1 | | Authors: | Stura, E.A, Vera, L, Maiga, A.A, Marchetti, C, Lorphelin, A, Bellanger, L, Servant, D, Gilles, N. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystallization of recombinant green mamba rho-Da1a toxin during a lyophilization procedure and its structure determination.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5QXO
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with DF848 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
3CU2
 
 | |
3G00
 
 | | Mth0212 in complex with a 9bp blunt end dsDNA at 1.7 Angstrom | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*GP*TP*AP*TP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*UP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
5QXZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with DF853 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
7EYO
 
 | | Crystal structure of leech hyaluronidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hyaluronoglucuronidase | | Authors: | Huang, H, Hou, X.D, Rao, Y.J, Kang, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and cleavage pattern of a hyaluronate 3-glycanohydrolase in the glycoside hydrolase 79 family.
Carbohydr Polym, 277, 2022
|
|
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
3PVG
 
 | | Crystal structure of Z. mays CK2 alpha subunit in complex with the inhibitor 4,5,6,7-tetrabromo-1-carboxymethylbenzimidazole (K68) | | Descriptor: | (4,5,6,7-tetrabromo-1H-benzimidazol-1-yl)acetic acid, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2010-12-07 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
