5WA9
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{R})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
8IE8
 
 | | Crystal structure of DAPK1 in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
6GSR
 
 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex at 5.5 Angstrom resolution. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P. | | Deposit date: | 2018-06-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
5WHO
 
 | |
6H40
 
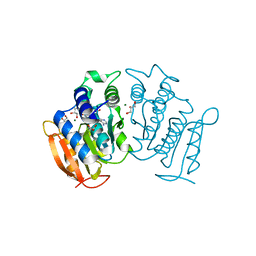 | | High resolution structure of MeT1 from Mycobacterium hassiacum in complex with 3-methoxy-1,2-propanediol. | | Descriptor: | (2~{S})-3-methoxypropane-1,2-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pereira, P.J.B, Ripoll-Rozada, J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.053 Å) | | Cite: | Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7MOG
 
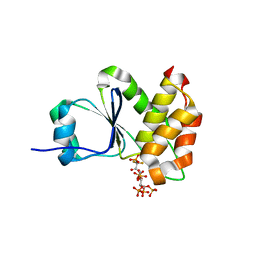 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-PCF2 Am-InsP5, an analogue of 5-InsP7 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
6H8J
 
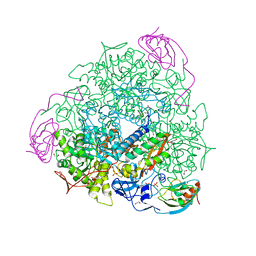 | | 1.45 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPTO | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-08-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into Urease Inhibition by N-( n-Butyl) Phosphoric Triamide through an Integrated Structural and Kinetic Approach.
J.Agric.Food Chem., 67, 2019
|
|
8AHY
 
 | |
8ALH
 
 | |
8ALM
 
 | |
8Q8I
 
 | | AO75L in complex with a synthetic trisaccharide acceptor. | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-methyl-6-octoxy-3-oxidanyl-oxan-4-yl]oxy-6-methyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, BICINE, ... | | Authors: | Laugueri, M.E, Speciale, I, Gimeno, A, Sicheng, L, Poveda, A, Lowary, T, Van Etten J, L, Barbero, J, De Castro, C, Tonetti, M, Rojas A, L. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Unveiling the GT114 family: Structural characterization of A075L, a glycosyltransferase from Paramecium bursaria chlorella virus-1 (PBCV-1).
Protein Sci., 33, 2024
|
|
6ZB9
 
 | | Exo-beta-1,3-glucanase from moose rumen microbiome, wild type | | Descriptor: | Exo-beta-1,3-glucanase | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
3ES4
 
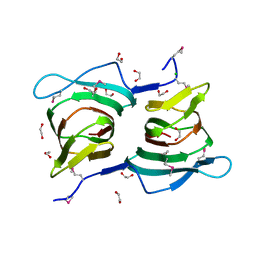 | |
6ZEA
 
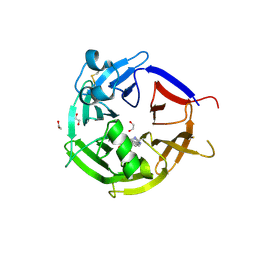 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
6P2G
 
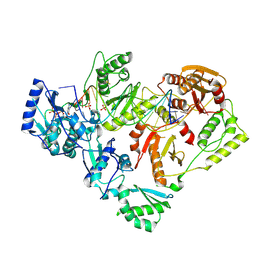 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and D-ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA Primer 20-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
5X79
 
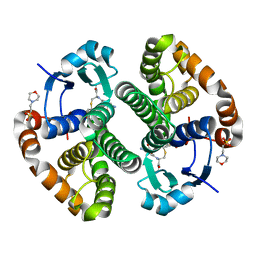 | | Human GST Pi conjugated with novel inhibitor, GS-ESF | | Descriptor: | (2S)-2-azanyl-5-[[(2R)-3-(2-fluorosulfonylethylsulfanyl)-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase P | | Authors: | Tomoike, F, Shishido, Y, Fukui, K, Kimura, Y, Abe, H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-09-13 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A covalent G-site inhibitor for glutathione S-transferase Pi (GSTP1-1).
Chem. Commun. (Camb.), 53, 2017
|
|
6OFI
 
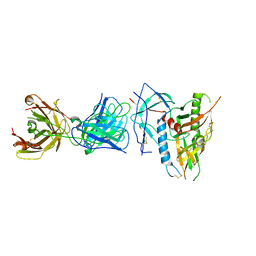 | | CRYSTAL STRUCTURE OF the RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH55 Fab heavy chain, CH55 Fab light chain, ... | | Authors: | Tolbert, W.D, Yan, F, Van, V, Pazgier, M. | | Deposit date: | 2019-03-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6NNN
 
 | |
7R4P
 
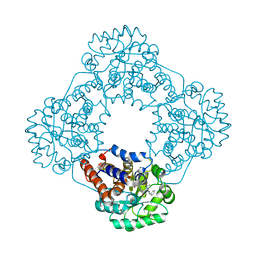 | | Structure of human hydroxyacid oxidase 1 bound with 6-amino-1-benzyl-5-(methylamino)pyrimidine-2,4(1H,3H)-dione | | Descriptor: | 6-amino-1-benzyl-5-(methylamino)pyrimidine-2,4(1H,3H)-dione, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-09 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
4YJL
 
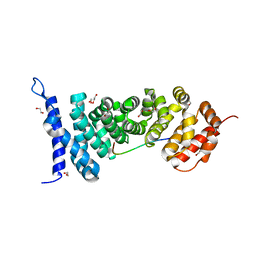 | | Crystal structure of APC-ARM in complexed with Amer1-A2 | | Descriptor: | 1,2-ETHANEDIOL, APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
6HP9
 
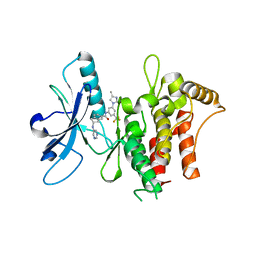 | | Structure of the kinase domain of human DDR1 in complex with a 2-Amino-2,3-Dihydro-1H-Indene-5-Carboxamide-based inhibitor | | Descriptor: | (2~{R})-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-2-(pyrimidin-5-ylamino)-2,3-dihydro-1~{H}-indene-5-carboxamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Amino-2,3-dihydro-1H-indene-5-carboxamide-Based Discoidin Domain Receptor 1 (DDR1) Inhibitors: Design, Synthesis, and in Vivo Antipancreatic Cancer Efficacy.
J.Med.Chem., 62, 2019
|
|
3FQ2
 
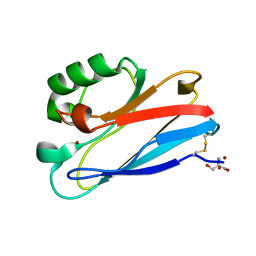 | | Azurin C112D/M121F | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Lancaster, K.M, Gray, H.B. | | Deposit date: | 2009-01-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Type Zero Copper Proteins.
Nat Chem, 1, 2009
|
|
9HN8
 
 | |
3DHB
 
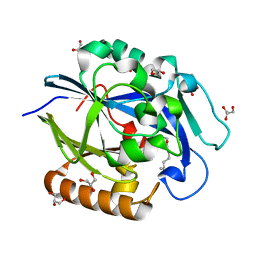 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
5L2N
 
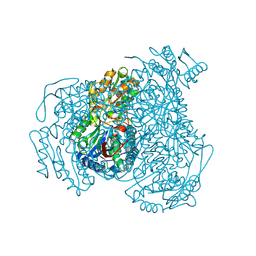 | | Structure of ALDH1A1 in complex with BUC25 | | Descriptor: | 3-benzyl-4-methyl-2-oxo-2H-1-benzopyran-7-yl methanesulfonate, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Buchman, C.D, Hurley, T.D. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives.
J. Med. Chem., 60, 2017
|
|
