6YLN
 
 | | mTurquoise2 SG P212121 - Directional optical properties of fluorescent proteins | | Descriptor: | EGFP, POTASSIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLQ
 
 | | EGFP in neutral pH, Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | GLYCEROL, MAGNESIUM ION, eGFP | | Authors: | Myskova, J, Rybakova, M, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5O9P
 
 | | Crystal structure of Gas2 in complex with compound 10 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(pyridin-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
2QD4
 
 | | Wild type human ferrochelatase crystallized with MnCl2 | | Descriptor: | CHLORIDE ION, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Medlock, A.E, Dailey, T.A, Ross, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2007-06-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pi-Helix Switch Selective for Porphyrin Deprotonation and Product Release in Human Ferrochelatase.
J.Mol.Biol., 373, 2007
|
|
1UO3
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
3CZO
 
 | |
2BC5
 
 | | Crystal structure of E. coli cytochrome b562 with engineered c-type heme linkages | | Descriptor: | HEME C, SULFATE ION, Soluble cytochrome b562 | | Authors: | Faraone-Mennella, J, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stability and Folding Kinetics of Structurally Characterized Cytochrome c-b(562).
Biochemistry, 45, 2006
|
|
6YJQ
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A,Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
1NIS
 
 | | CRYSTAL STRUCTURE OF ACONITASE WITH TRANS-ACONITATE AND NITROCITRATE BOUND | | Descriptor: | 2-HYDROXY-2-NITROMETHYL SUCCINIC ACID, ACONITASE, IRON/SULFUR CLUSTER | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1993-01-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of aconitase with trans-aconitate and nitrocitrate bound.
J.Mol.Biol., 237, 1994
|
|
4DK7
 
 | | Crystal structure of LXR ligand binding domain in complex with full agonist 1 | | Descriptor: | ACETATE ION, CALCIUM ION, N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-methylbenzenesulfonamide, ... | | Authors: | Piper, D.E, Xu, H. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of a new binding mode for a series of liver X receptor agonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1N84
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
4KQ8
 
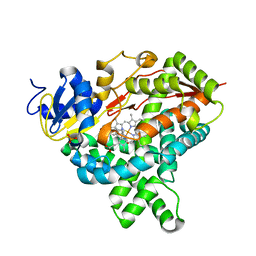 | | Structure of Recombinant Human Cytochrome P450 Aromatase | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Cytochrome P450 19A1, PHOSPHATE ION, ... | | Authors: | Ghosh, D, Di Nardo, G, Griswold, J. | | Deposit date: | 2013-05-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the functional roles of critical residues in human cytochrome p450 aromatase.
Biochemistry, 52, 2013
|
|
1CSV
 
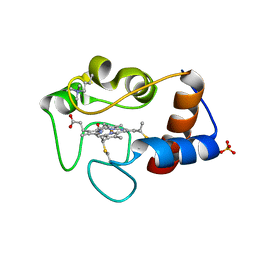 | |
1CSU
 
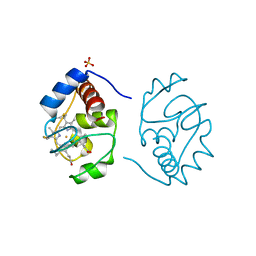 | |
1CSX
 
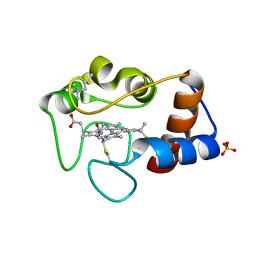 | |
1CSW
 
 | |
6LFH
 
 | |
1NEG
 
 | | Crystal Structure Analysis of N-and C-terminal labeled SH3-domain of alpha-Chicken Spectrin | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Mueller, U, Buessow, K, Diehl, A, Niesen, F.H, Nyarsik, L, Heinemann, U. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid purification and crystal structure analysis of a small protein carrying two terminal affinity tags
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
4K9G
 
 | | 1.55 A Crystal Structure of Macrophage Migration Inhibitory Factor bound to ISO-66 and a related compound | | Descriptor: | (4R,6Z)-6-(3-fluoro-4-hydroxyphenyl)-4-hydroxy-6-iminohexan-2-one, 1-[(5S)-3-(3-fluoro-4-hydroxyphenyl)-4,5-dihydro-1,2-oxazol-5-yl]propan-2-one, CHLORIDE ION, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ISO-66, a novel inhibitor of macrophage migration, shows efficacy in melanoma and colon cancer models.
INT J ONCOL., 45, 2014
|
|
1QNG
 
 | |
2P4Q
 
 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
6IEP
 
 | | GAPDH of Streptococcus agalactiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Ponnuraj, K, Nagarajan, R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GAPDH of Streptococcus agalactiae and characterization of its interaction with extracellular matrix molecules
Microb. Pathog., 127, 2018
|
|
5NUK
 
 | | Engineered beta-lactoglobulin: variant I56F-L39A-M107F in complex with chlorpromazine (LG-FAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin, CHLORIDE ION, ... | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Lazinska, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
232L
 
 | | T4 LYSOZYME MUTANT M120K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
1E1C
 
 | | METHYLMALONYL-COA MUTASE H244A Mutant | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, METHYLMALONYL-COA MUTASE ALPHA CHAIN, ... | | Authors: | Evans, P.R, Thoma, N.H. | | Deposit date: | 2000-04-30 | | Release date: | 2000-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Protection of Radical Intermediates at the Active Site of Adenosylcobalamin-Dependent Methylmalonyl-Coa Mutase
Biochemistry, 39, 2000
|
|
