7PD9
 
 | |
6AMP
 
 | | Crystal structure of H172A PHM (CuH absent, CuM present) | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
4P8R
 
 | |
7PCP
 
 | |
4PCN
 
 | | Phosphotriesterase variant R22 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R22, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-04-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
4L4B
 
 | | Structure of L358A/K178G/D182N mutant of P450cam bound to camphor | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
7PJY
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in chimeric state 1 (CHI1-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
5XMJ
 
 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
4PCV
 
 | | The structure of BdcA (YjgI) from E. coli | | Descriptor: | BdcA (YjgI), PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Page, R, Peti, W, Lord, D. | | Deposit date: | 2014-04-16 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BdcA, a protein important for Escherichia coli biofilm dispersal, is a short-chain dehydrogenase/reductase that binds specifically to NADPH.
Plos One, 9, 2014
|
|
7PJX
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in hybrid state 1 (H1-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6GTA
 
 | | Alpha-galactosidase mutant D378A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 3,5 difluorophenyl leaving group | | Descriptor: | (1~{R},2~{S},3~{S},6~{S})-6-[3,5-bis(fluoranyl)phenoxy]-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
5XGV
 
 | |
4PD2
 
 | | Crystal structure of a complex between a C248GH LlFpg mutant and a THF containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Zinc finger oxidation of Fpg/Nei DNA glycosylases by 2-thioxanthine: biochemical and X-ray structural characterization.
Nucleic Acids Res., 42, 2014
|
|
2W8P
 
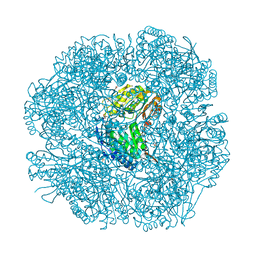 | | The crystal structure of human C340A SSADH | | Descriptor: | GLYCEROL, SUCCINIC SEMIALDEHYDE DEHYDROGENASE MITOCHONDRIAL, SULFATE ION | | Authors: | Kim, Y.-G, Kim, K.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-Switch Modulation of Human Ssadh by Dynamic Catalytic Loop.
Embo J., 28, 2009
|
|
4L4G
 
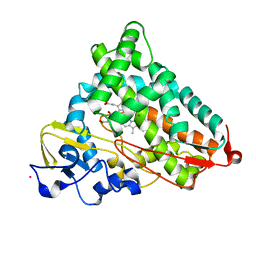 | | Structure of cyanide and camphor bound P450cam mutant L358P/K178G | | Descriptor: | CAMPHOR, CYANIDE ION, Camphor 5-monooxygenase, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
6G6P
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.2 in P63 | | Descriptor: | Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
4L4P
 
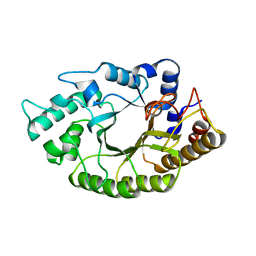 | | the mutant(E139A) structure in complex with xylotriose | | Descriptor: | Endo-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | An, J, Feng, Y, Wu, G. | | Deposit date: | 2013-06-08 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of CbXyn10B from Caldicellulosiruptor bescii and its mutant(E139A) in complex with xylotriose
To be Published
|
|
6GMF
 
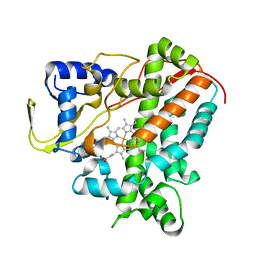 | | Structure of Cytochrome P450 CYP109Q5 from Chondromyces apiculatus | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Grogan, G, Dubiel, P, Sharma, M, Klenk, J, Hauer, B. | | Deposit date: | 2018-05-25 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization and structure-guided engineering of the novel versatile terpene monooxygenase CYP109Q5 from Chondromyces apiculatus DSM436.
Microb Biotechnol, 12, 2019
|
|
7PJZ
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in chimeric state 2 (CHI2-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6AOO
 
 | | 2.15 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae | | Descriptor: | Malate dehydrogenase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae.
To Be Published
|
|
5XNL
 
 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
7PJW
 
 | | Structure of the 70S-EF-G-GDP-Pi ribosome complex with tRNAs in hybrid state 2 (H2-EF-G-GDP-Pi) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6AP6
 
 | | Crystal Structure of DAD2 in complex with tolfenamic acid | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, Probable strigolactone esterase DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of strigolactone receptors byN-phenylanthranilic acid derivatives: Structural and functional insights.
J. Biol. Chem., 293, 2018
|
|
7PLE
 
 | | ArsH of Paracoccus denitrificans | | Descriptor: | NADPH-dependent FMN reductase | | Authors: | Kryl, M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The ArsH Protein Product of the Paracoccus denitrificans ars Operon Has an Activity of Organoarsenic Reductase and Is Regulated by a Redox-Responsive Repressor.
Antioxidants, 11, 2022
|
|
6G7O
 
 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
