7EU5
 
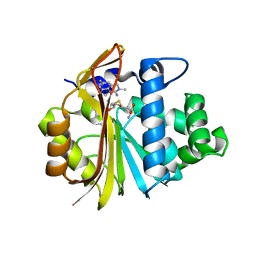 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000107 | | Descriptor: | 6-fluoranyl-10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-16 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
1O6Z
 
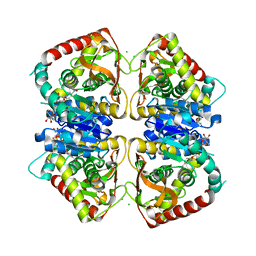 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
3NEA
 
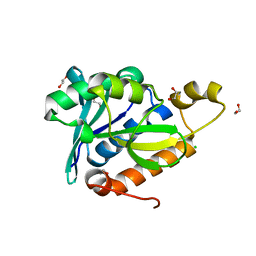 | | Crystal Structure of Peptidyl-tRNA hydrolase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Lam, R, McGrath, T.E, Romanov, V, Gothe, S.A, Peddi, S.R, Razumova, E, Lipman, R.S, Branstrom, A.A, Chirgadze, N.Y. | | Deposit date: | 2010-06-08 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Francisella tularensis peptidyl-tRNA hydrolase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2HP3
 
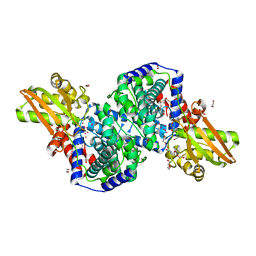 | | Crystal structure of iminodisuccinate epimerase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IDS-epimerase, ... | | Authors: | Lohkamp, B, Bauerle, B, Rieger, P.G, Schneider, G. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Three-dimensional Structure of Iminodisuccinate Epimerase Defines the Fold of the MmgE/PrpD Protein Family.
J.Mol.Biol., 362, 2006
|
|
9FQ5
 
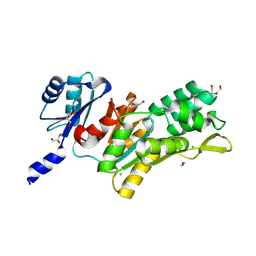 | |
5KQV
 
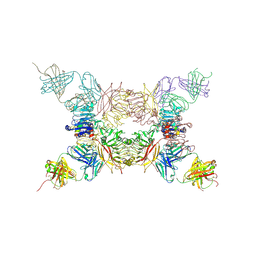 | | Insulin receptor ectodomain construct comprising domains L1,CR,L2, FnIII-1 and alphaCT peptide in complex with bovine insulin and FAB 83-14 (REVISED STRUCTURE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, Insulin receptor,Insulin receptor, ... | | Authors: | Lawrence, M.C, Smith, B.J, Croll, T.I. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor.
Nature, 493, 2013
|
|
4OQF
 
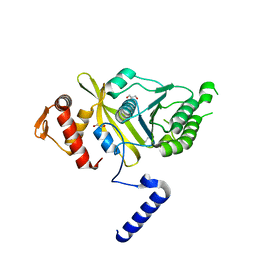 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIB-SR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
3TIY
 
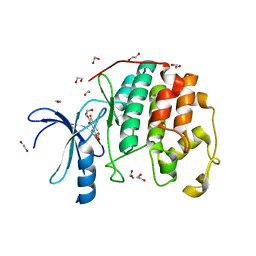 | | CDK2 in complex with NSC 35676 | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
1H41
 
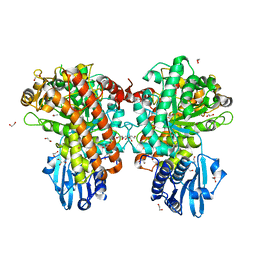 | | Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid, ALPHA-GLUCURONIDASE, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-09-25 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Alpha-Glucuronidase,Glca67A,of Cellvibrio Japonicus Utilizes the Carboxylate and Methyl Groups of Aldobiouronic Acid as Important Substrate Recognition Determinants
J.Biol.Chem., 278, 2003
|
|
4DFE
 
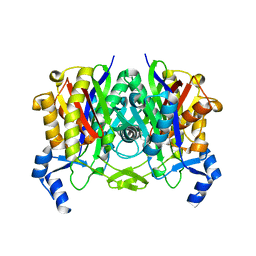 | |
2XOL
 
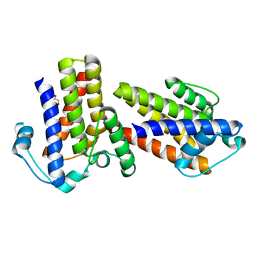 | | High resolution structure of TtrD from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHAPERONE PROTEIN TTRD | | Authors: | Dawson, A, Coulthurst, S.J, Sargent, F, Hunter, W.N. | | Deposit date: | 2010-08-18 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conserved Signal Peptide Recognition Systems Across the Prokaryotic Domains.
Biochemistry, 51, 2012
|
|
3CXS
 
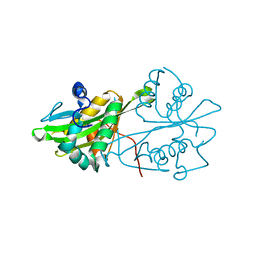 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
5L66
 
 | |
4CS2
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in its apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PYRROLYSINE--TRNA LIGASE | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
6HA9
 
 | |
4CS4
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, 2-{[dihydroxy(4-aminoethylphenyl)-{4}-sulfanyl]amino}-3-hydroxypropanoic acid, MAGNESIUM ION, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
4OA5
 
 | |
7HTT
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z56960248 | | Descriptor: | 1-[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]-N-methylmethanamine, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2STD
 
 | | SCYTALONE DEHYDRATASE COMPLEXED WITH TIGHT-BINDING INHIBITOR CARPROPAMID | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, SCYTALONE DEHYDRATASE, SULFATE ION | | Authors: | Nakasako, M, Motoyama, T, Kurahashi, Y, Yamaguchi, I. | | Deposit date: | 1997-12-21 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cryogenic X-ray crystal structure analysis for the complex of scytalone dehydratase of a rice blast fungus and its tight-binding inhibitor, carpropamid: the structural basis of tight-binding inhibition.
Biochemistry, 37, 1998
|
|
1YQ7
 
 | | Human farnesyl diphosphate synthase complexed with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION, ... | | Authors: | Kavanagh, K.L, Guo, K, Von delft, F, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human farnesyl diphosphate complexed with clinical inhibitor risedronate
To be Published
|
|
5T79
 
 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | Descriptor: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
7HT5
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z32400357 | | Descriptor: | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3SPB
 
 | | Unliganded E. Cloacae MurA | | Descriptor: | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-07-01 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
1O4Z
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF BETA-AGARASE B FROM ZOBELLIA GALACTANIVORANS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Allouch, J, Jam, M, Helbert, W, Barbeyron, T, Kloareg, B, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Two {beta}-Agarases.
J.Biol.Chem., 278, 2003
|
|
