7K6O
 
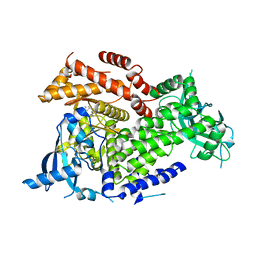 | | Crystal structure of PI3Kalpha inhibitor 10-5429 | | Descriptor: | (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-methylpyrrolidine-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
6B3E
 
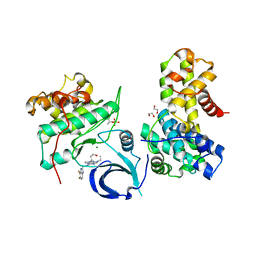 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
2V5J
 
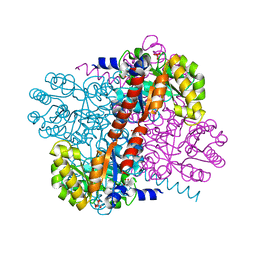 | | Apo Class II aldolase HpcH | | Descriptor: | 2,4-DIHYDROXYHEPT-2-ENE-1,7-DIOIC ACID ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Rea, D, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2007-07-05 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of Hpch: A Metal Ion Dependent Class II Aldolase from the Homoprotocatechuate Degradation Pathway of Escherichia Coli.
J.Mol.Biol., 373, 2007
|
|
5TV3
 
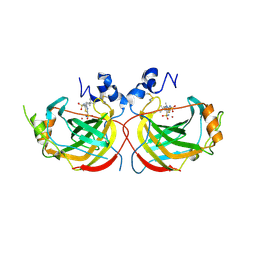 | | Crystal structure of the complex of Helicobacter pylori alpha-carbonic anhydrase with (E)-5-(((4-(tert-butyl)phenyl)sulfonyl)imino)-4-methyl-4,5-dihydro-1,3,4-thiadiazole-2-sulfonamide | | Descriptor: | (5Z)-5-{[(4-tert-butylphenyl)sulfonyl]imino}-4-methyl-4,5-dihydro-1,3,4-thiadiazole-2-sulfonamide, Alpha-carbonic anhydrase, CHLORIDE ION, ... | | Authors: | Modak, J.K, Roujeinikova, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Activity Relationship for Sulfonamide Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase.
J. Med. Chem., 59, 2016
|
|
4PO8
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-CR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4ZAZ
 
 | | Structure of UbiX Y169F in complex with a covalent adduct formed between reduced FMN and dimethylallyl monophosphate | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
7K6N
 
 | | Crystal structure of PI3Kalpha selective Inhibitor 11-1575 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, tert-butyl (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-3-methylpyrrolidine-1-carboxylate | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
8ZWP
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
5VB2
 
 | |
4P9T
 
 | | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin | | Descriptor: | 1,2-ETHANEDIOL, Catenin alpha-2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shibahara, T, Hirano, Y, Hakoshima, T. | | Deposit date: | 2014-04-04 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin.
Febs Lett., 589, 2015
|
|
1OBW
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Oganessyan, V.Yu, Harutyunyan, E.H, Avaeva, S.M, Oganessyan, N.N, Mather, T, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Biochemistry, 36, 1997
|
|
3G7N
 
 | | Crystal Structure of a Triacylglycerol Lipase from Penicillium Expansum at 1.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase, PENTAETHYLENE GLYCOL, ... | | Authors: | Bian, C.B, Yuan, C, Chen, L.Q, Edward, J.M, Lin, L, Jiang, L.G, Huang, Z.X, Huang, M.D. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD
Proteins, 78, 2010
|
|
8ZWW
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
1D11
 
 | | INTERACTIONS BETWEEN AN ANTHRACYCLINE ANTIBIOTIC AND DNA MOLECULAR STRUCTURE OF DAUNOMYCIN COMPLEXED TO D(CPGPTPAPCPG) AT 1.2-ANGSTROMS RESOLUTION | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), SODIUM ION | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Rich, A. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-A resolution.
Biochemistry, 26, 1987
|
|
2QIS
 
 | | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Kavanagh, K.L, Dunford, J.E, Hozjan, V, Evdokimov, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate.
To be Published
|
|
5EDQ
 
 | | EGFR kinase (T790M/L858R) with inhibitor compound 15: ~{N}-(7-chloranyl-1~{H}-indazol-3-yl)-7,7-dimethyl-2-(1~{H}-pyrazol-4-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Descriptor: | Epidermal growth factor receptor, ~{N}-(7-chloranyl-1~{H}-indazol-3-yl)-7,7-dimethyl-2-(1~{H}-pyrazol-4-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6V74
 
 | | Crystal Structure of Human PKM2 in Complex with L-asparagine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ASPARAGINE, CHLORIDE ION, ... | | Authors: | Nandi, S, Dey, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Biochemical and structural insights into how amino acids regulate pyruvate kinase muscle isoform 2.
J.Biol.Chem., 295, 2020
|
|
5IXT
 
 | |
5DR3
 
 | | Endothiapepsin in complex with fragment 333 | | Descriptor: | 1,2-ETHANEDIOL, 4-propan-2-ylsulfanyl-1-propyl-6,7-dihydro-5~{H}-cyclopenta[d]pyrimidin-2-one, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
1COT
 
 | |
5DRY
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [N-(1-(2-chlorophenyl)-1H-indol-6-yl)-2-(2-(5-(2-chlorophenyl)-1H-tetrazol-1-yl)acetyl)hydrazinecarboxamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-[1-(2-chlorophenyl)-1H-indol-6-yl]-2-{[5-(2-chlorophenyl)-1H-tetrazol-1-yl]acetyl}hydrazinecarboxamide, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5VCL
 
 | | Structure of the Qdm peptide bound to Qa-1a | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H2-T23 protein, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Qa-1a with bound Qa-1 determinant modifier peptide.
PLoS ONE, 12, 2017
|
|
5SNS
 
 | |
4PP5
 
 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 5-methyl UAB30 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-(5-methyl-3,4-dihydronaphthalen-1(2H)-ylidene)octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity.
J.Med.Chem., 57, 2014
|
|
4M8X
 
 | | GS-8374, a Novel Phosphonate-Containing Inhibitor of HIV-1 Protease, Effectively Inhibits HIV PR Mutants with Amino Acid Insertions | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Grantz saskova, K, Brynda, J, Rezacova, P, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-08-14 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GS-8374, a prototype phosphonate-containing inhibitor of HIV-1 protease, effectively inhibits protease mutants with amino acid insertions.
J.Virol., 88, 2014
|
|
