5O84
 
 | | Glutathione S-transferase Tau 23 (partially oxidized) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Young, D.R, Van Molle, I, Tossounian, M, Messens, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
7AEJ
 
 | |
6TZY
 
 | |
6U0D
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
5O8Y
 
 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5O6G
 
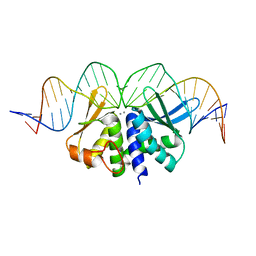 | |
5O9H
 
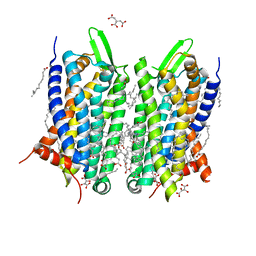 | | Crystal structure of thermostabilised human C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, C5a anaphylatoxin chemotactic receptor 1, CITRIC ACID, ... | | Authors: | Robertson, N, Rappas, M, Dore, A.S, Brown, J, Bottegoni, G, Koglin, M, Cansfield, J, Jazayeri, A, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727.
Nature, 553, 2018
|
|
6UI4
 
 | | Crystal structure of phenamacril-bound F. graminearum myosin I | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, Y, Zhou, X.E, Gong, Y, Zhu, Y, Xu, H.E, Zhou, M, Melcher, K, Zhang, F. | | Deposit date: | 2019-09-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Fusarium myosin I inhibition by phenamacril.
Plos Pathog., 16, 2020
|
|
6UIM
 
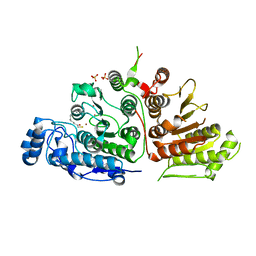 | |
5OCF
 
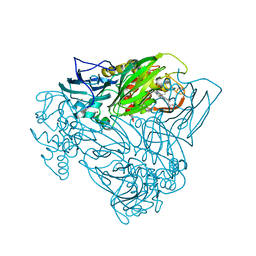 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCP
 
 | |
6U2X
 
 | | Structure of ALDH7A1 mutant E399G complexed with NAD | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6U3U
 
 | | Crystal Structure of Shiga Toxin 2K | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Shiga toxin 2K subunit A, ... | | Authors: | Zhang, Y.Z, He, X.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2.
Microorganisms, 8, 2019
|
|
5OG0
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5OGE
 
 | |
6UKU
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 3 | | Descriptor: | 4,4'-[propane-1,3-diylbis(6-methoxy-1-benzothiene-5,2-diyl)]bis(4-oxobutanoic acid), fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
5OA0
 
 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, W164F) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, STRYCHNINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
7B4O
 
 | |
5OHV
 
 | | K33-specific affimer bound to K33 diUb | | Descriptor: | GLYCEROL, K33-specific affimer, SULFATE ION, ... | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5OAN
 
 | | Crystal structure of mutant AChBP in complex with glycine (T53F, Q74R, Y110A, I135S, G162E, S206CCP_KGTG) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-23 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
6ULZ
 
 | | Adenylation domain of the initiation module of LgrA mutant P483M | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FORMIC ACID, ... | | Authors: | Chiche-Lapierre, C, Alonzo, D.A, Schmeing, T.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis.
Nat.Chem.Biol., 16, 2020
|
|
5OAV
 
 | |
7B4P
 
 | |
5OKC
 
 | |
6U7A
 
 | | Rv3722c in complex with kynurenine | | Descriptor: | (2S)-4-(2-aminophenyl)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-hydroxyquinoline-2-carboxylic acid, ... | | Authors: | Mandyoli, L, Sacchettini, J. | | Deposit date: | 2019-09-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
