6ABI
 
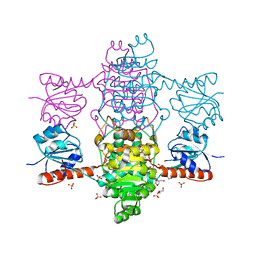 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6ABJ
 
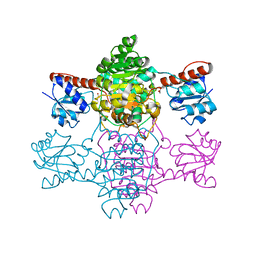 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
7XAY
 
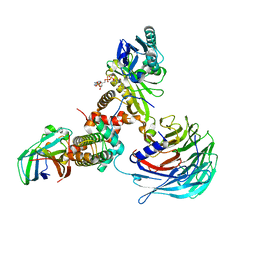 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
6HY2
 
 | |
4EJX
 
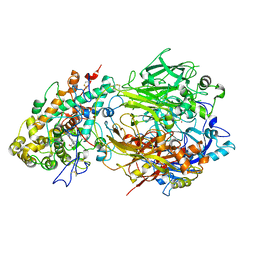 | | Structure of ceruloplasmin-myeloperoxidase complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Ceruloplasmin, ... | | Authors: | Samygina, V.R, Sokolov, A.V, Bourenkov, G, Vasilyev, V.B, Bartunik, H. | | Deposit date: | 2012-04-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.69 Å) | | Cite: | Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins.
Plos One, 8, 2013
|
|
9BDP
 
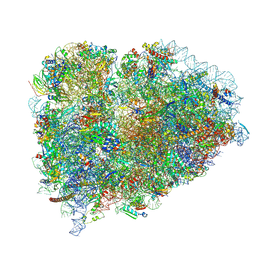 | |
6EBN
 
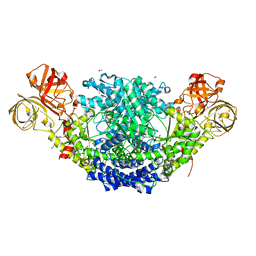 | | Crystal structure of Psilocybe cubensis noncanonical aromatic amino acid decarboxylase | | Descriptor: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Torrens-Spence, M.P, Chun-Ting, L, Pluskal, T, Chung, Y.K, Weng, J.K. | | Deposit date: | 2018-08-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9663111 Å) | | Cite: | Monoamine Biosynthesis via a Noncanonical Calcium-Activatable Aromatic Amino Acid Decarboxylase in Psilocybin Mushroom.
ACS Chem. Biol., 13, 2018
|
|
4R4Q
 
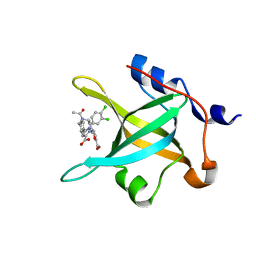 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
6EH0
 
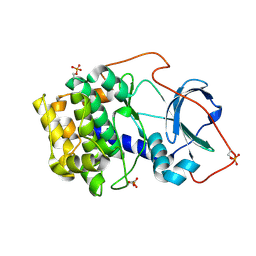 | |
7OPD
 
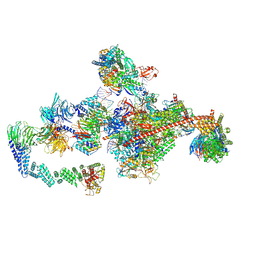 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 5) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
9CA2
 
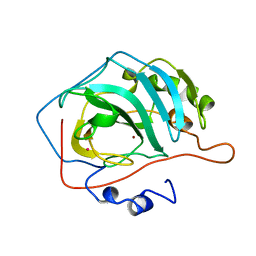 | |
7OPC
 
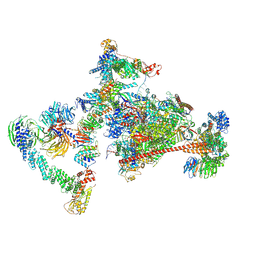 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 4) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
6E4W
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-(4,6-difluoro-5-{4-[(2S)-oxan-2-yl]phenyl}-1H-indole-3-carbonyl)-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
3KWB
 
 | | Structure of CatK covalently bound to a dioxo-triazine inhibitor | | Descriptor: | 3,5-dioxo-4-(3-piperidin-1-ylpropyl)-2-[3-(trifluoromethyl)phenyl]-2,3,4,5-tetrahydro-1,2,4-triazine-6-carbonitrile, Cathepsin K | | Authors: | Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Dioxo-triazines as a novel series of cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KW9
 
 | |
3KWZ
 
 | |
6E5M
 
 | | Crystallographic structure of the cyclic nonapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 32 2 1 | | Descriptor: | 9MER-PEPTIDE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Crystallographic structure of a complex between trypsin and a nonapeptide derived from a Bowman-Birk inhibitor found in Vigna unguiculata seeds.
Arch. Biochem. Biophys., 665, 2019
|
|
4R4C
 
 | | Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)-2-chlorobenzamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({[4-(5-carboxyfuran-2-yl)-2-chlorobenzoyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
6E8L
 
 | | Crystal Structure of Alkyl hydroperoxidase D (AhpD) from Streptococcus pneumoniae (Strain D39/ NCTC 7466) | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Meng, Y, Davies, J, North, R, Coombes, D, Horne, C, Hampton, M, Dobson, R. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function analyses of alkylhydroperoxidase D fromStreptococcus pneumoniaereveal an unusual three-cysteine active site architecture.
J.Biol.Chem., 295, 2020
|
|
5TF9
 
 | |
6ESA
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp120 and AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, beta-D-ribopyranose, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
6X1Y
 
 | | Mre11 dimer in complex with small molecule modulator PFMI | | Descriptor: | (5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclease SbcCD subunit D | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6ERW
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp013 and Fasudil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, UPF0418 protein FAM164A, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
6E5K
 
 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, Aib helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, Aib helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
5TI9
 
 | | Crystal structure of human TDO in complex with Trp and dioxygen, Northeast Structural Genomics Consortium Target HR6161 | | Descriptor: | N'-Formylkynurenine, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Forouhar, F, Lewis-Ballester, A, Lew, S, Karkashon, S, Seetharaman, J, Yeh, S.R, Tong, L. | | Deposit date: | 2016-10-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for catalysis and substrate-mediated cellular stabilization of human tryptophan 2,3-dioxygenase.
Sci Rep, 6, 2016
|
|
