7XZY
 
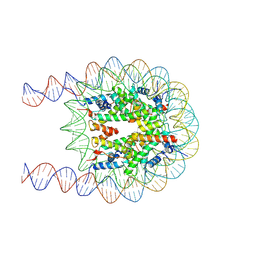 | | Cryo-EM structure of the nucleosome containing 193 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZX
 
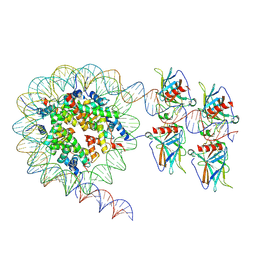 | | Cryo-EM structure of the nucleosome in complex with p53 DNA-binding domain | | Descriptor: | Cellular tumor antigen p53, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
6N6W
 
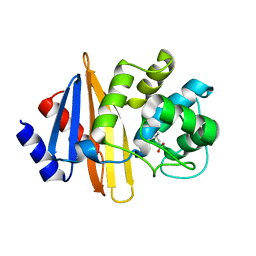 | | OXA-23 mutant F110A/M221A neutral pH form | | Descriptor: | Beta-lactamase oxa23 | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D beta-Lactamases.
Antimicrob. Agents Chemother., 63, 2019
|
|
7XSP
 
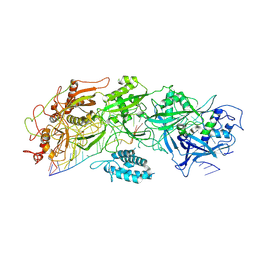 | | Structure of gRAMP-target RNA | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSR
 
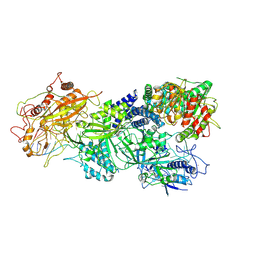 | | Structure of Craspase-target RNA | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSQ
 
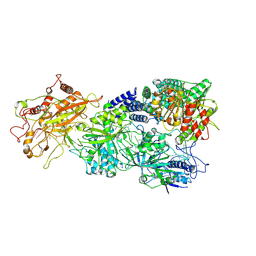 | | Structure of the Craspase | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSS
 
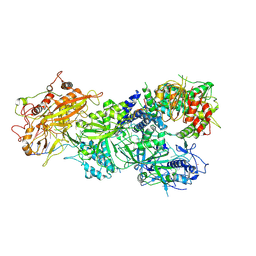 | | Structure of Craspase-CTR | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
8F2W
 
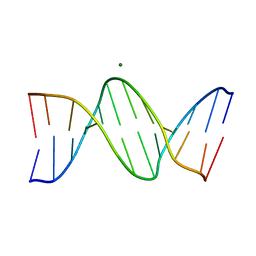 | | Structure of a B-Form Dodecamer: 5'-CGCGAATTCGCG-3 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-08 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
6N6V
 
 | |
6I0V
 
 | | Crystal structure of DmTailor in complex with CACAGU RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*AP*CP*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
6N6T
 
 | | OXA-23 mutant F110A/M221A low pH form | | Descriptor: | Beta-lactamase oxa23, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D beta-Lactamases.
Antimicrob. Agents Chemother., 63, 2019
|
|
8PRX
 
 | | Crystal structure of human cathepsin L after reaction with the bound ketoamide inhibitor 13b | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-07-12 | | Release date: | 2023-08-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6N6Y
 
 | |
8EZR
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
8EZT
 
 | | Crystal structure of HipB(Lp) from Legionella pneumophila | | Descriptor: | CHLORIDE ION, HipB(Lp) | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of HipB(Lp) from Legionella pneumophila
To Be Published
|
|
8EZS
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein | | Descriptor: | CHLORIDE ION, HipS(Lp), HipT(Lp) | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein
To Be Published
|
|
3QSY
 
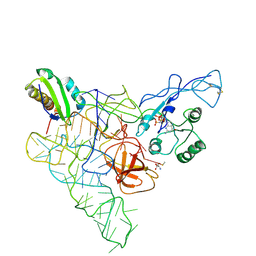 | | Recognition of the methionylated initiator tRNA by the translation initiation factor 2 in Archaea | | Descriptor: | METHIONINE, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Translation initiation factor 2 subunit alpha, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Zelinskaya, N.V, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2011-02-22 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the archaeal translation initiation factor 2 in complex with a GTP analogue and Met-tRNAf(Met.)
J.Mol.Biol., 425, 2013
|
|
4ITB
 
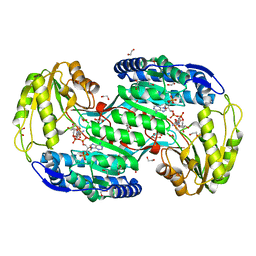 | | Structure of bacterial enzyme in complex with cofactor and substrate | | Descriptor: | 1,2-ETHANEDIOL, 4-oxobutanoic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rhee, S, Park, J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-24 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
8ENX
 
 | |
8ENY
 
 | |
8EO0
 
 | |
8ENZ
 
 | |
8ENS
 
 | |
8ENW
 
 | |
8EFZ
 
 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
