8G7Q
 
 | |
6EZJ
 
 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8GIZ
 
 | | E. coli clamp loader with open clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
6EXN
 
 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
8GLT
 
 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
8GIY
 
 | | E. coli clamp loader with closed clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8G7R
 
 | |
8G7P
 
 | |
6F2P
 
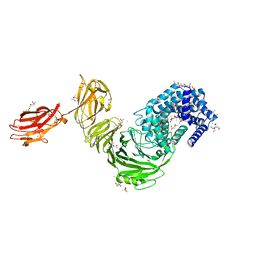 | | Structure of Paenibacillus xanthan lyase to 2.6 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lo Leggio, L, Kadziola, A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Dynamics of a Promiscuous Xanthan Lyase from Paenibacillus nanensis and the Design of Variants with Increased Stability and Activity.
Cell Chem Biol, 26, 2019
|
|
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | Descriptor: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | Authors: | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
8G7S
 
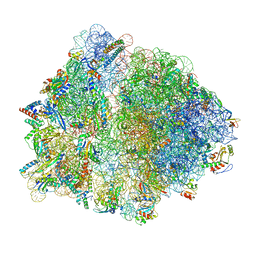 | |
6EZ0
 
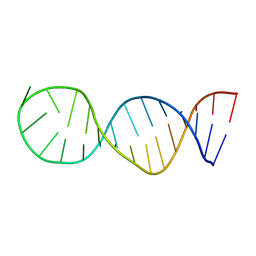 | | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding | | Descriptor: | RNA (27-MER) | | Authors: | Erat, M.C, Besic, E, Oberhuber, M, Johannsen, S, Sigel, R.K.O. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding.
J. Biol. Inorg. Chem., 23, 2018
|
|
8GI6
 
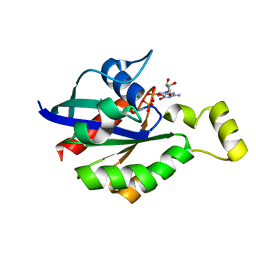 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
6F2H
 
 | | Structure of Protease 1 from Pyrococcus horikoshii co-crystallized in presence of 10 mM Tb-Xo4 and potassium iodide. | | Descriptor: | Deglycase PH1704, IODIDE ION, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Franzetti, B, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6F7U
 
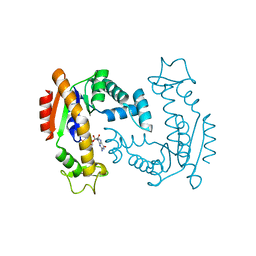 | | Molecular Mechanism of ATP versus GTP Selectivity of Adenylate Kinase | | Descriptor: | Adenylate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Rogne, P, Rosselin, M, Grundstrom, C, Hedberg, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2017-12-12 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of ATP versus GTP selectivity of adenylate kinase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8FZD
 
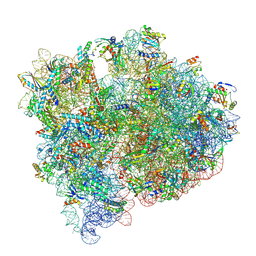 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZE
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZI
 
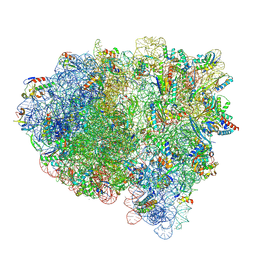 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZF
 
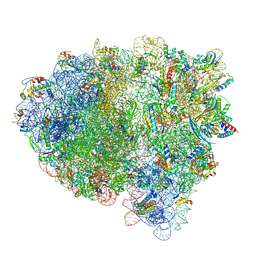 | | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZG
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZJ
 
 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZH
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8G4I
 
 | | 40S ribosomal subunit of the 80S Giardia intestinalis assemblage A ribosome with Emetine bound in V1 conformation | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S25, ... | | Authors: | Eiler, D.R, Wimberly, B.T, Bilodeau, D.Y, Rissland, O.S, Kieft, J.S. | | Deposit date: | 2023-02-09 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The Giardia lamblia ribosome structure reveals divergence in several biological pathways and the mode of emetine function.
Structure, 32, 2024
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
