1JKZ
 
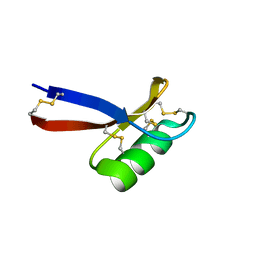 | | NMR Solution Structure of Pisum sativum defensin 1 (Psd1) | | Descriptor: | DEFENSE-RELATED PEPTIDE 1 | | Authors: | Almeida, M.S, Cabral, K.M.S, Kurtenbach, E, Almeida, F.C.L, Valente, A.P. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action.
J.Mol.Biol., 315, 2002
|
|
1KUH
 
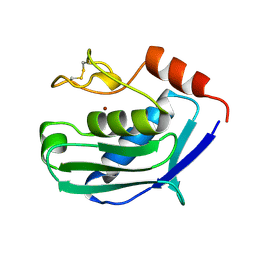 | | ZINC PROTEASE FROM STREPTOMYCES CAESPITOSUS | | Descriptor: | CALCIUM ION, ZINC ION, ZINC PROTEASE | | Authors: | Kurisu, G, Kinoshita, T, Sugimoto, A, Nagara, A, Kai, Y, Kasai, N, Harada, S. | | Deposit date: | 1996-02-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the zinc endoprotease from Streptomyces caespitosus.
J.Biochem.(Tokyo), 121, 1997
|
|
1LBQ
 
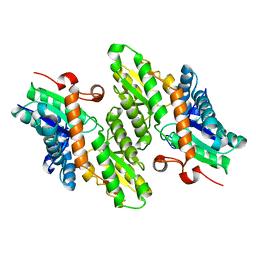 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
1JBK
 
 | |
1K48
 
 | |
1KUI
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEQW. | | Descriptor: | CADMIUM ION, EQW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KQH
 
 | | NMR Solution Structure of the cis Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-05 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
1KQI
 
 | | NMR Solution Structure of the trans Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
1LA8
 
 | | Solution structure of the DNA hairpin 13-mer CGCGGTGTCCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*TP*GP*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-28 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the 1,N(2)-propanodeoxyguanosine adduct in a three-base DNA hairpin loop derived from a palindrome in the Salmonella typhimurium hisD3052 gene.
Chem.Res.Toxicol., 15, 2002
|
|
1LRQ
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP, A5P and Cadmium | | Descriptor: | ARABINOSE-5-PHOSPHATE, CADMIUM ION, KDO-8-phosphate synthetase, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
1JEC
 
 | |
1JF4
 
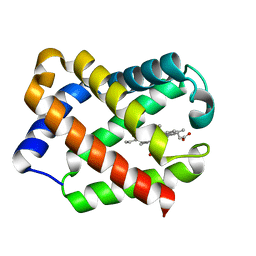 | | Crystal Structure Of Component IV Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component IV | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1JJZ
 
 | |
1JJ9
 
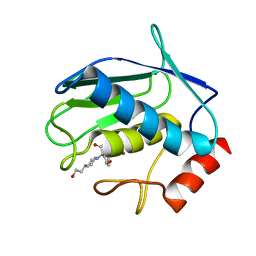 | | Crystal Structure of MMP8-Barbiturate Complex Reveals Mechanism for Collagen Substrate Recognition | | Descriptor: | 2-HYDROXY-5-[4-(2-HYDROXY-ETHYL)-PIPERIDIN-1-YL]-5-PHENYL-1H-PYRIMIDINE-4,6-DIONE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Brandstetter, H, Grams, F, Glitz, D, Lang, A, Huber, R, Bode, W, Krell, H.-W, Engh, R.A. | | Deposit date: | 2001-07-04 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8-A crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition.
J.Biol.Chem., 276, 2001
|
|
1JEE
 
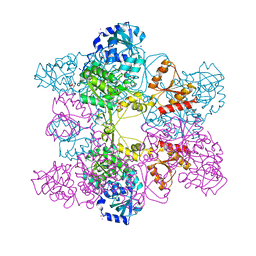 | | Crystal Structure of ATP Sulfurylase in complex with chlorate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JME
 
 | | Crystal Structure of Phe393His Cytochrome P450 BM3 | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ost, T.W.B, Munro, A.W, Mowat, C.G, Pesseguiero, A, Fulco, A.J, Cho, A.K, Cheesman, M.A, Walkinshaw, M.D, Chapman, S.K. | | Deposit date: | 2001-07-18 | | Release date: | 2001-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and spectroscopic analysis of the F393H mutant of flavocytochrome P450 BM3.
Biochemistry, 40, 2001
|
|
1JWE
 
 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
1JZC
 
 | |
1MV3
 
 | |
1MUZ
 
 | |
1IVH
 
 | | STRUCTURE OF HUMAN ISOVALERYL-COA DEHYDROGENASE AT 2.6 ANGSTROMS RESOLUTION: STRUCTURAL BASIS FOR SUBSTRATE SPECIFICITY | | Descriptor: | COENZYME A PERSULFIDE, FLAVIN-ADENINE DINUCLEOTIDE, ISOVALERYL-COA DEHYDROGENASE | | Authors: | Tiffany, K.A, Roberts, D.L, Wang, M, Paschke, R, Mohsen, A.-W.A, Vockley, J, Kim, J.J.P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity,.
Biochemistry, 36, 1997
|
|
1N4E
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA decamer containing a cis-syn thymine dimer.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MOZ
 
 | | ADP-ribosylation factor-like 1 (ARL1) from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-like protein 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Amor, J.C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structures of Yeast ARF2 and ARL1:
DISTINCT ROLES FOR THE N TERMINUS IN THE STRUCTURE
AND FUNCTION OF ARF FAMILY GTPases
J.Biol.Chem., 276, 2001
|
|
1MXC
 
 | | S-ADENOSYLMETHIONINE SYNTHETASE WITH 8-BR-ADP | | Descriptor: | 8-BROMOADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Markham, G.D. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of S-adenosylmethionine synthetase: crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution.
Biochemistry, 35, 1996
|
|
