1RLC
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
2UXZ
 
 | | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary- Alcohol-Containing Transition-State Mimic | | Descriptor: | HIV-1 PROTEASE, METHYL [(1S)-1-({2-[(4R)-4-BENZYL-4-HYDROXY-5-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-5-OXOPENTYL]-2-(4-BROMOBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE | | Authors: | Ginman, N, Samuelsson, B, Hallberg, A, Unge, J.T, Unge, T.K. | | Deposit date: | 2007-04-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary-Alcohol-Containing Transition-State Mimic.
J.Med.Chem., 51, 2008
|
|
2UY0
 
 | | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary- Alcohol-Containing Transition-State Mimic | | Descriptor: | HIV-1 PROTEASE, METHYL [(1S)-1-({2-[(4R)-4-BENZYL-4-HYDROXY-5-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-5-OXOPENTANOYL]-2-(4-BROMOBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE | | Authors: | Ginman, N, Samuelsson, B, Hallberg, A, Unge, J.T, Unge, T.K. | | Deposit date: | 2007-04-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary-Alcohol-Containing Transition-State Mimic.
J.Med.Chem., 51, 2008
|
|
2BE2
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with R221239 | | Descriptor: | 4-(3,5-DIMETHYLPHENOXY)-5-(FURAN-2-YLMETHYLSULFANYLMETHYL)-3-IODO-6-METHYLPYRIDIN-2(1H)-ONE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Himmel, D.M, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Nguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
1B6P
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 7 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL) -PROPYL]-3-METHYL-2-PROPIONYLAMINO-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1BP8
 
 | | 4:2:1 mithramycin:Mg++:d(ACCCGGGT)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-METHYL-ANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, 5'-D(*AP*CP*CP*CP*GP*GP*GP*T)-3', ... | | Authors: | Keniry, M.A, Owen, E.A, Shafer, R.H. | | Deposit date: | 1998-08-13 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the 4:1 mithramycin:d(ACCCGGGT)2 complex: evidence for an interaction between the E saccharides
Biopolymers, 54, 2000
|
|
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
2WON
 
 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (wild-type). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lersivirine: A Non-Nucleoside Reverse Transcriptase Inhibitor with Activity Against Drug- Resistant Human Immunodeficiency Virus-1.
Antimicrob.Agents Chemother., 54, 2010
|
|
4P9H
 
 | |
1RPK
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
3HR9
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase F40I mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | Authors: | Tsai, L.C, Huang, H.C, Hsiao, C.H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase mutant F40I
To be Published
|
|
3RPL
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
2WOM
 
 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (K103N). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1.
Antimicrob. Agents Chemother., 54, 2010
|
|
2FHX
 
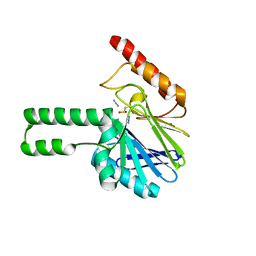 | | Pseudomonas aeruginosa SPM-1 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Murphy, T.A, Catto, L.E, Halford, S.E, Hadfield, A.T, Minor, W, Walsh, T.R, Spencer, J. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pseudomonas aeruginosa SPM-1 Provides Insights into Variable Zinc Affinity of Metallo-beta-lactamases.
J.Mol.Biol., 357, 2006
|
|
4EEG
 
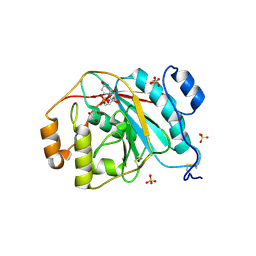 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-Gal-Beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EEA
 
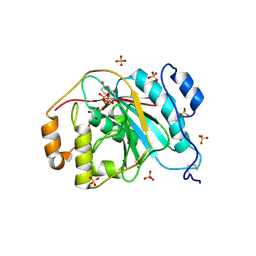 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-Gal-Beta1,4-Glc-BETA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
1ST8
 
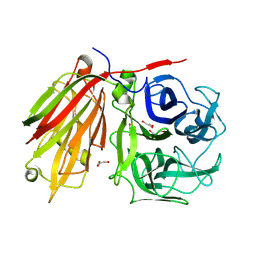 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Van den Ende, W, De Ranter, C.J, Van Laere, A, Rabijns, A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus.
Plant J., 41, 2005
|
|
3R0Y
 
 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors | | Descriptor: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2S)-1-{[(2S,3S)-3-hydroxy-5-oxo-5-{[(2R)-1-oxo-3-phenyl-1-(prop-2-yn-1-ylamino)propan-2-yl]amino}-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | Authors: | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | Deposit date: | 2011-03-09 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
4EE3
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with pentasaccharide | | Descriptor: | 6-AMINOHEXYL-URIDINE-C1,5'-DIPHOSPHATE, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
2CJT
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
3TT0
 
 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
3H0O
 
 | | The importance of CH-Pi stacking interactions between carbohydrate and aromatic residues in truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | Authors: | Tsai, L.C, Hsiao, C.H. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The importance of CH-Pi stacking interactions between carbohydrate and aromatic residues in truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
To be Published
|
|
1B6K
 
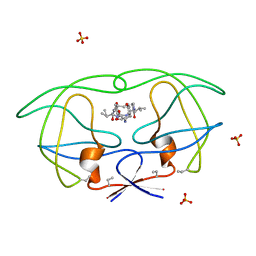 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 5 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2]HEPTADECA-1(16),13(17),14- TRIEN-11-YLAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL)-PROPYL]-3-METHYL-2- (2-OXO-PYRROLIDIN-1-YL)-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
2HRQ
 
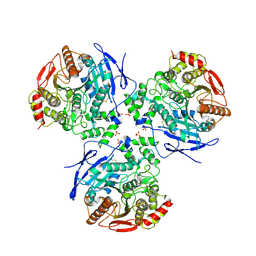 | | Crystal structure of Human Liver Carboxylesterase 1 (hCE1) in covalent complex with the nerve agent Soman (GD) | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (R)-METHYLPHOSPHINATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Liver carboxylesterase 1, ... | | Authors: | Fleming, C.D, Redinbo, M.R. | | Deposit date: | 2006-07-20 | | Release date: | 2007-05-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human Carboxylesterase 1 in Covalent Complexes with the Chemical Warfare Agents Soman and Tabun.
Biochemistry, 46, 2007
|
|
1AJK
 
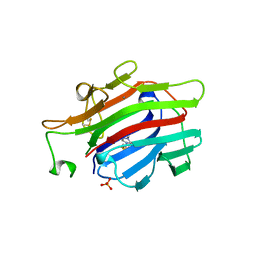 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
