1C4O
 
 | | CRYSTAL STRUCTURE OF THE DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB FROM THERMUS THERMOPHILUS | | Descriptor: | DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Machius, M, Henry, L, Palnitkar, M, Deisenhofer, J. | | Deposit date: | 1999-09-14 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C4P
 
 | | BETA DOMAIN OF STREPTOKINASE | | Descriptor: | PROTEIN (STREPTOKINASE) | | Authors: | Wang, X, Zhang, X.C. | | Deposit date: | 1999-09-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of streptokinase beta-domain.
FEBS Lett., 459, 1999
|
|
1C4Q
 
 | |
1C4R
 
 | | THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REGULATION OF LNS DOMAIN FUNCTION BY ALTERNATIVE SPLICING | | Descriptor: | NEUREXIN-I BETA | | Authors: | Rudenko, G, Nguyen, T, Chelliah, Y, Sudhof, T.C, Deisenhofer, J. | | Deposit date: | 1999-09-28 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing.
Cell(Cambridge,Mass.), 99, 1999
|
|
1C4S
 
 | | CHONDROITIN-4-SULFATE. THE STRUCTURE OF A SULFATED GLYCOSAMINOGLYCAN | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-4-deoxy-beta-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Chondroitin 4-sulfate: the structure of a sulfated glycosaminoglycan.
J.Mol.Biol., 125, 1978
|
|
1C4T
 
 | | CATALYTIC DOMAIN FROM TRIMERIC DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | PROTEIN (DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE), SULFATE ION | | Authors: | Knapp, J.E, Carroll, D, Lawson, J.E, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1999-09-22 | | Release date: | 2000-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expression, purification, and structural analysis of the trimeric form of the catalytic domain of the Escherichia coli dihydrolipoamide succinyltransferase.
Protein Sci., 9, 2000
|
|
1C4U
 
 | | SELECTIVE NON ELECTROPHILIC THROMBIN INHIBITORS WITH CYCLOHEXYL MOIETIES. | | Descriptor: | 2-[2-(4-BROMO-BENZENESULFONYL)-ETHYL]-1-3-DIOXO-2,3,5,8-TETRAHYDRO-1H-[1,2,4]TRIAZOLO[1,2-A]PYRIDAZINE-5-CARBOXYLIC ACID(4-CARBAMIMIDOYL-CYCLOHEXYLMETHYL)-AMIDE, PROTEIN (HIRUGEN), SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R.K, Tulinsky, A. | | Deposit date: | 1999-09-25 | | Release date: | 2000-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C4V
 
 | | SELECTIVE NON ELECTROPHILIC THROMBIN INHIBITORS WITH CYCLOHEXYL MOIETIES. | | Descriptor: | 2-(2,2-DIPHENYL-ETHYL)-7-METHYL-1,3-DIOXO-2,3,5,8-TETRAHYDRO-1H-[1,2,4]TRIAZOLO [1,2-A]PYRIDAZINE-5-CARBOXYLIC ACID(4-CARBAMIMIDOYL-CYCLOHEXYLMETHYL)-AMIDE, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R.K, Tulinsky, A. | | Deposit date: | 1999-09-25 | | Release date: | 2000-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C4W
 
 | | 1.9 A STRUCTURE OF A-THIOPHOSPHONATE MODIFIED CHEY D57C | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Halkides, C.J, McEvoy, M.M, Matsumura, P, Volz, K, Dahlquist, F.W. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY.
Biochemistry, 39, 2000
|
|
1C4X
 
 | | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE (BPHD) FROM RHODOCOCCUS SP. STRAIN RHA1 | | Descriptor: | PROTEIN (2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE) | | Authors: | Nandhagopal, N, Senda, T, Mitsui, Y. | | Deposit date: | 1999-09-30 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic Acid (Hpda) Hydrolase (Bphd Enzyme) from Rhodococcus Sp. Strain Rha1, in the Pcb Degradation Pathway
Proc.Jpn.Acad.,Ser.B, 73, 1997
|
|
1C4Y
 
 | | SELECTIVE NON-ELECTROPHILIC THROMBIN INHIBITORS | | Descriptor: | 2-(2,2-DIPHENYL-ETHYL)-7-METHYL-1,3-DIOXO-2,3,5,8-TETRAHYDRO-1H-[1,2,4] TRIAZOLO[1,2-A]PYRIDAZINE-5-CARBOXYLIC ACID [4-(2-AMINO-3H-IMIDAZOL-4-YL)-CYCLOHEXYL]-AMIDE, HIRUGEN, THROMBIN:LONG CHAIN, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R.K, Tulinsky, A. | | Deposit date: | 1999-10-05 | | Release date: | 2000-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C4Z
 
 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1C50
 
 | | IDENTIFICATION AND STRUCTURAL CHARACTERIZATION OF A NOVEL ALLOSTERIC BINDING SITE OF GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Skamnaki, V.T, Tsitsanou, K.E, Gavalas, N.G, Johnson, L.N. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new allosteric site in glycogen phosphorylase b as a target for drug interactions.
Structure Fold.Des., 8, 2000
|
|
1C51
 
 | | PHOTOSYNTHETIC REACTION CENTER AND CORE ANTENNA SYSTEM (TRIMERIC), ALPHA CARBON ONLY | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Klukas, O, Schubert, W.D, Jordan, P, Krauss, N, Fromme, P, Witt, H.T, Saenger, W. | | Deposit date: | 1999-10-21 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Photosystem I, an improved model of the stromal subunits PsaC, PsaD, and PsaE.
J.Biol.Chem., 274, 1999
|
|
1C52
 
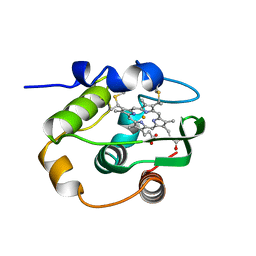 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1C53
 
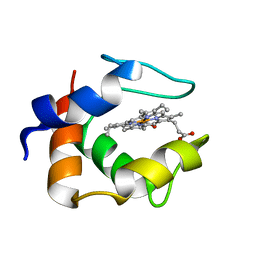 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
1C54
 
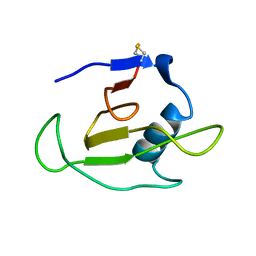 | | SOLUTION STRUCTURE OF RIBONUCLEASE SA | | Descriptor: | RIBONUCLEASE SA | | Authors: | Laurents, D.V, Canadillas-Perez, J.M, Santoro, J, Schell, D, Pace, C.N, Rico, M, Bruix, M. | | Deposit date: | 1999-10-22 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ribonuclease Sa.
Proteins, 44, 2001
|
|
1C55
 
 | |
1C56
 
 | |
1C57
 
 | | DIRECT DETERMINATION OF THE POSITIONS OF DEUTERIUM ATOMS OF BOUND WATER IN CONCANAVALIN A BY NEUTRON LAUE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, Concanavalin-Br, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb, A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C58
 
 | | CRYSTAL STRUCTURE OF CYCLOAMYLOSE 26 | | Descriptor: | Cyclohexacosakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Gessler, K, Saenger, W, Nimz, O. | | Deposit date: | 1999-11-04 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | V-Amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose).
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C5A
 
 | |
1C5B
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C5C
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX | | Descriptor: | 2-ACETYLAMINO-NAPTHALENE-1,5-DISULFONIC ACID, CHIMERIC DECARBOXYLASE ANTIBODY 21D8, GLYCEROL | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-09 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C5D
 
 | | THE CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A RAT MONOCLONAL ANTIBODY AGAINST THE MAIN IMMUNOGENIC REGION OF THE HUMAN MUSCLE ACETYLCHOLINE RECEPTOR | | Descriptor: | MONOCLONAL ANTIBODY AGAINST THE MAIN IMMUNOGENIC REGION OF THE HUMAN MUSCLE ACETYLCHOLINE RECEPTOR | | Authors: | Kontou, M, Leonidas, D.D, Vatzaki, E.H, Tsantili, P, Mamalaki, A, Oikonomakos, N.G, Acharya, K.R, Tzartos, S.J. | | Deposit date: | 1999-11-17 | | Release date: | 1999-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the Fab fragment of a rat monoclonal antibody against the main immunogenic region of the human muscle acetylcholine receptor.
Eur.J.Biochem., 267, 2000
|
|
