7ZEE
 
 | | Human cytosolic 5' nucleotidase IIIB | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methylguanosine phosphate-specific 5'-nucleotidase, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
7G0N
 
 | | Crystal Structure of delipidated apo human FABP4 | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1LNI
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS AT ATOMIC RESOLUTION (1.0 A) | | Descriptor: | GLYCEROL, GUANYL-SPECIFIC RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution data reveal flexibility in the structure of RNase Sa.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4NAE
 
 | | PcrB from Geobacillus kaustophilus, with bound G1P | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Heptaprenylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Peterhoff, D, Beer, B, Rajendran, C, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-10-22 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
6UAZ
 
 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with glucose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
5L6K
 
 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-30 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
1POB
 
 | | CRYSTAL STRUCTURE OF COBRA-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE | | Descriptor: | 1-O-OCTYL-2-HEPTYLPHOSPHONYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | White, S.P, Scott, D.L, Otwinowski, Z, Sigler, P.B. | | Deposit date: | 1992-09-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue.
Science, 250, 1990
|
|
5GK6
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Citrate bound form | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Citrate bound form
To Be Published
|
|
3MC4
 
 | |
5VMK
 
 | |
7ZI0
 
 | | Structure of human Smoothened in complex with cholesterol and SAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-N-[trans-4-(methylamino)cyclohexyl]-N-{[3-(pyridin-4-yl)phenyl]methyl}-1-benzothiophene-2-carboxamide, CHOLESTEROL, ... | | Authors: | Byrne, E.F.X, Woolley, R.E, Ansell, B, Sansom, M.S.P, Newstead, S, Siebold, C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Patched 1 regulates Smoothened by controlling sterol binding to its extracellular cysteine-rich domain.
Sci Adv, 8, 2022
|
|
5LHV
 
 | | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with uridine and sulfate ion at 1.29 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Balaev, V.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-07-13 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with uridine and sulfate ion at 1.29 A resolution
To Be Published
|
|
6DOG
 
 | |
5LLH
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-(1,3-Benzothiazol-2-ylthio)-2,3,5,6-tetrafluorobenzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(1,3-benzothiazol-2-ylsulfanyl)-2,3,5,6-tetrakis(fluoranyl)benzenesulfonamide, BICINE, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
5F13
 
 | | Structure of Mn bound DUF89 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nocek, B, Skarina, T, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | A family of metal-dependent phosphatases implicated in metabolite damage-control.
Nat.Chem.Biol., 12, 2016
|
|
5LSZ
 
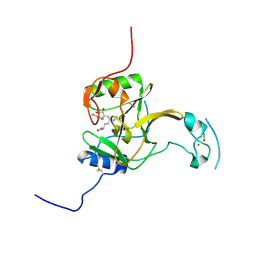 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
6DPD
 
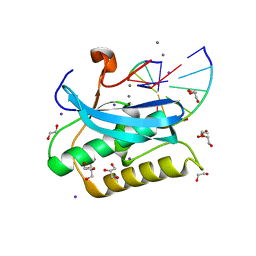 | |
2WEI
 
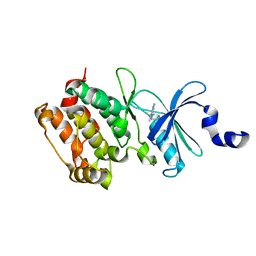 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
5CT0
 
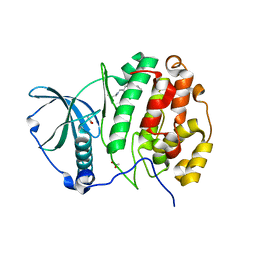 | | Crystal structure of CK2alpha with 3-(3-chloro-4-(phenyl)benzylamino)propan-1-ol bound | | Descriptor: | 3-{[(2-chlorobiphenyl-4-yl)methyl]amino}propan-1-ol, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
4BCR
 
 | | Structure of PPARalpha in complex with WY14643 | | Descriptor: | 1,2-ETHANEDIOL, 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR ALPHA | | Authors: | Bernardes, A, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2012-10-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Mechanism of Peroxisome Proliferator-Activated Receptor Alpha Activation by Wy14643: A New Mode of Ligand Recognition and Receptor Stabilization
J.Mol.Biol., 425, 2013
|
|
5D2F
 
 | | 4-oxalocrotonate decarboxylase from Pseudomonas putida G7 - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-oxalocrotonate decarboxylase NahK, ACETATE ION, ... | | Authors: | Guimaraes, S.L, Nagem, R.A.P. | | Deposit date: | 2015-08-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Crystal Structures of Apo and Liganded 4-Oxalocrotonate Decarboxylase Uncover a Structural Basis for the Metal-Assisted Decarboxylation of a Vinylogous beta-Keto Acid.
Biochemistry, 55, 2016
|
|
6TW8
 
 | | Leishmania major N-myristoyltransferase in complex with indazole inhibitor IMP-917 | | Descriptor: | 1-[5-[4-fluoranyl-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-2~{H}-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2020-01-12 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Leishmania major N-myristoyltransferase in complex with indazole inhibitor IMP-917
To Be Published
|
|
5D3Z
 
 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase in complex with a small phosphonate inhibitor | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
3CH9
 
 | |
6DOB
 
 | |
