7SCV
 
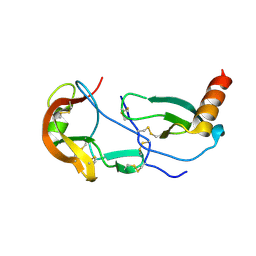 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCT
 
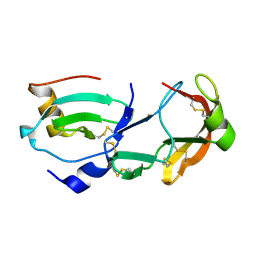 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
6VWA
 
 | |
2KHZ
 
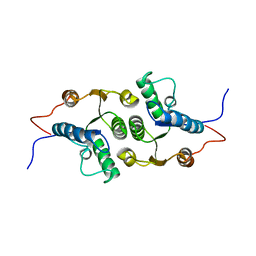 | | Solution Structure of RCL | | Descriptor: | c-Myc-responsive protein Rcl | | Authors: | Doddapaneni, K, Mahler, B, Yuan, C, Wu, Z. | | Deposit date: | 2009-04-15 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RCL, a novel 2'-deoxyribonucleoside 5'-monophosphate N-glycosidase
J.Mol.Biol., 394, 2009
|
|
2CBH
 
 | |
6CWS
 
 | |
6NJX
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with mercury | | Descriptor: | IODIDE ION, MERCURY (II) ION, Xcc_ctr_Hg | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
1AA9
 
 | | HUMAN C-HA-RAS(1-171)(DOT)GDP, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-HA-RAS, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ito, Y, Yamasaki, Y, Muto, Y, Kawai, G, Nishimura, S, Miyazawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-27 | | Release date: | 1997-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein.
Biochemistry, 36, 1997
|
|
6NJW
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with platinum | | Descriptor: | IODIDE ION, MAGNESIUM ION, PLATINUM (II) ION, ... | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NJV
 
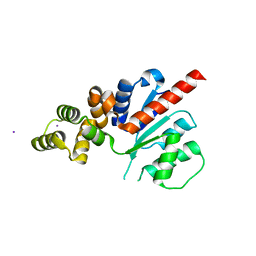 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with iodine | | Descriptor: | IODIDE ION, MAGNESIUM ION, Xcc_CTR_I | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
5OXI
 
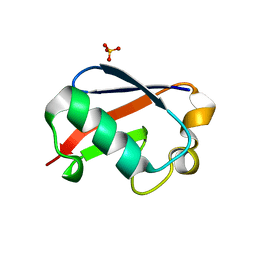 | | C-terminally retracted ubiquitin L67S mutant | | Descriptor: | SULFATE ION, Ubiquitin L67S mutant | | Authors: | Gladkova, C.G, Schubert, A.F, Wagstaff, J.L, Pruneda, J.N, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
5M58
 
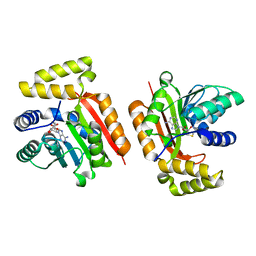 | |
1SOP
 
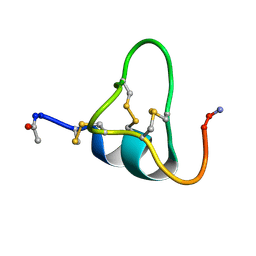 | |
6NK8
 
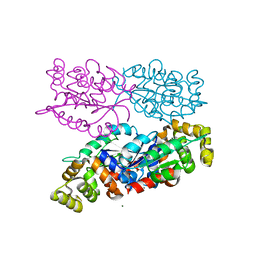 | | C-terminal region of the Burkholderia pseudomallei OLD protein | | Descriptor: | Class 2 OLD family nuclease, MAGNESIUM ION | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
8OSE
 
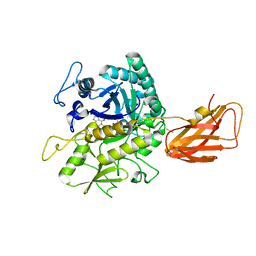 | | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), 1,2-ETHANEDIOL, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
6ONS
 
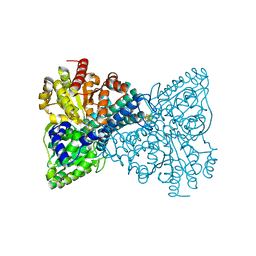 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase with the D-cluster ligating cysteines mutated to alanines, coexpressed with CooC, as-isolated | | Descriptor: | Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Cohen, S.E, Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
8DX1
 
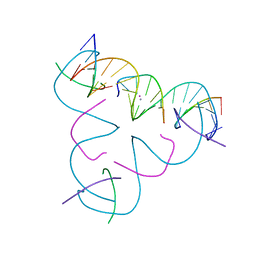 | | [C:Hg2+/Ag+:C--pH 7 MOPS] Metal-mediated DNA base pair in tensegrity triangle in Ag+ and Hg2+ solution in MOPS | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*CP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.79 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
6NOA
 
 | |
6CTD
 
 | |
3EGN
 
 | |
4BML
 
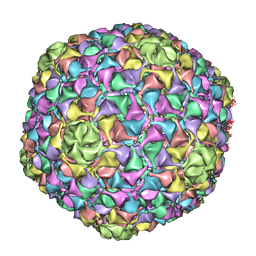 | | C-alpha backbone trace of major capsid protein gp39 found in marine virus Syn5. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Gipson, P, Baker, M.L, Raytcheva, D, Haase-Pettingell, C, Piret, J, King, J, Chiu, W. | | Deposit date: | 2013-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Protruding Knob-Like Proteins Violate Local Symmetries in an Icosahedral Marine Virus.
Nat.Commun., 5, 2014
|
|
6VW9
 
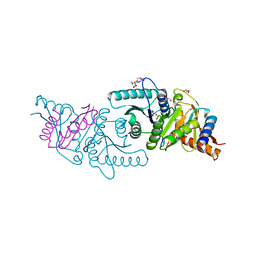 | | C-terminal regulatory domain of the chloride transporter KCC-1 from C. elegans, proteolyzed during crystallization | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, K+/Cl-Cotransporter | | Authors: | Zimanyi, C.M, Cheung, J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Regulatory Cytosolic Domain of a Eukaryotic Potassium-Chloride Cotransporter.
Structure, 28, 2020
|
|
5GIM
 
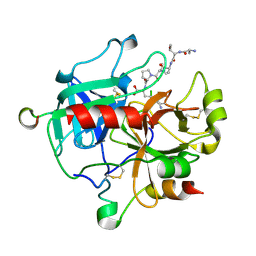 | | Crystal structure of thrombin-avathrin complex | | Descriptor: | C-terminal peptide from Putative uncharacterized protein avahiru, N-terminal peptide from Putative uncharacterized protein avahiru, Thrombin light chain, ... | | Authors: | Kini, R.M, Koh, C.Y, Iyer, J.K, Swaminathan, K. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Avathrin: a novel thrombin inhibitor derived from a multicopy precursor in the salivary glands of the ixodid tick, Amblyomma variegatum.
FASEB J., 31, 2017
|
|
1JUN
 
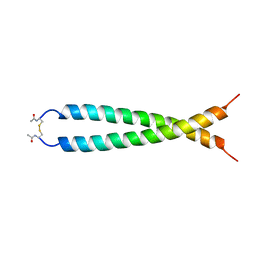 | |
2YHF
 
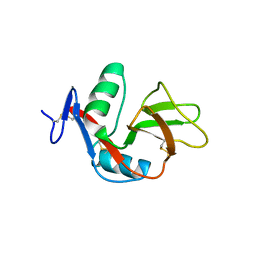 | | 1.9 Angstrom Crystal Structure of CLEC5A | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 5 MEMBER A | | Authors: | Watson, A.A, Lebedev, A.A, Murshudov, G.M, Vagin, A.A, Hall, B.A, O'Callaghan, C.A. | | Deposit date: | 2011-04-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Flexibility of the Macrophage Dengue Virus Receptor Clec5A: Implications for Ligand Binding and Signaling.
J.Biol.Chem., 286, 2011
|
|
