1HFI
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
4RUX
 
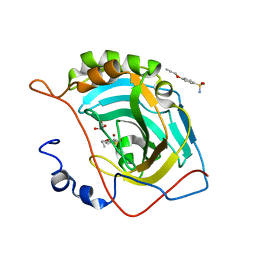 | |
1N32
 
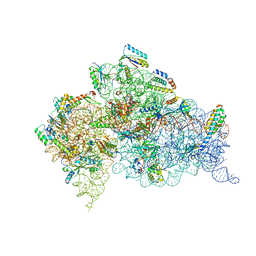 | | Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position at the a site with paromomycin | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Murphy IV, F.V, Tarry, M.J, Ramakrishnan, V. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-29 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form
Cell(Cambridge,Mass.), 111, 2002
|
|
4ZOT
 
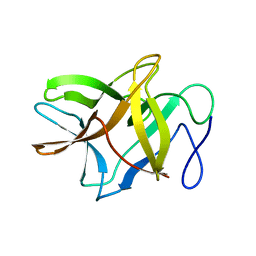 | | Crystal structure of BbKI, a disulfide-free plasma kallikrein inhibitor at 1.4 A resolution | | Descriptor: | Kunitz-type serine protease inhibitor BbKI | | Authors: | Shabalin, I.G, Zhou, D, Wlodawer, A, Oliva, M.L.V. | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of BbKI, a disulfide-free plasma kallikrein inhibitor.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1HXW
 
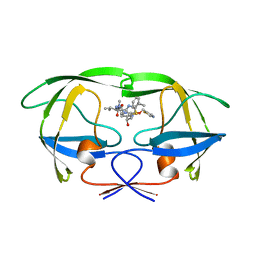 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-84538 | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Park, C.H, Nienaber, V, Kong, X.P. | | Deposit date: | 1997-01-24 | | Release date: | 1998-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
5E5F
 
 | |
5AO9
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-native | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5HPF
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain from Acinetobacter | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
5AGY
 
 | | CRYSTAL STRUCTURE OF A TAU CLASS GST MUTANT FROM GLYCINE | | Descriptor: | 4-NITROPHENYL METHANETHIOL, GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, ... | | Authors: | Axarli, I, Muleta, A.W, Vlachakis, D, Kossida, S, Kotzia, G, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2015-02-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Directed Evolution of Tau Class Glutathione Transferases Reveals a Site that Regulates Catalytic Efficiency and Masks Cooperativity.
Biochem.J., 473, 2016
|
|
5ACC
 
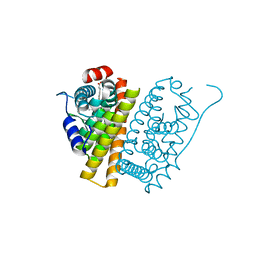 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | Descriptor: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
6EGO
 
 | |
5AOB
 
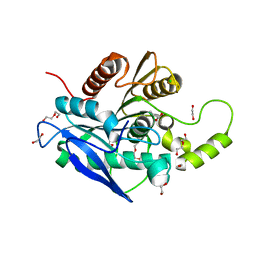 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AOC
 
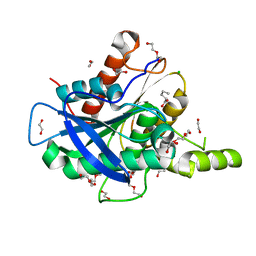 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AOA
 
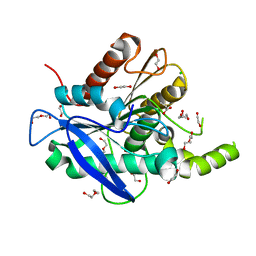 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
7BNT
 
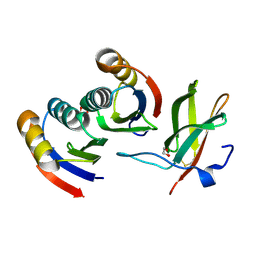 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
7BW2
 
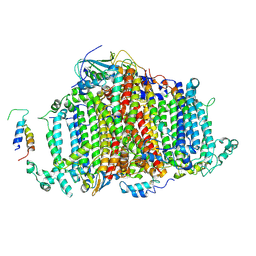 | | Crystal Structure of Cyanobacterial PSI Monomer from T.elongatus at 6.5 A Resolution | | Descriptor: | Photosystem I 4.8K protein, Photosystem I P700 chlorophyll a apoprotein A1, Photosystem I P700 chlorophyll a apoprotein A2, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Eithar, E.M, Mian, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
7JP2
 
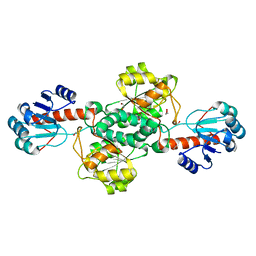 | | Crystal structure of TP0037 from Treponema pallidum, a D-lactate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-lactate dehydrogenase | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biophysical and Biochemical Characterization of TP0037, a d-Lactate Dehydrogenase, Supports an Acetogenic Energy Conservation Pathway in Treponema pallidum.
Mbio, 11, 2020
|
|
4YE1
 
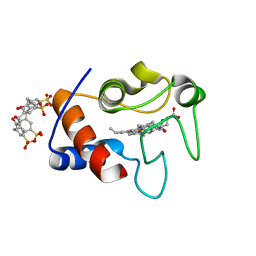 | | A cytochrome c plus calixarene structure - alternative ligand binding mode | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, GLYCEROL, ... | | Authors: | Mallon, M.M, McGovern, R.E, McCarty, A.A, Crowley, P.B. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A cytochrome c-calixarene structure
To Be Published
|
|
5E5E
 
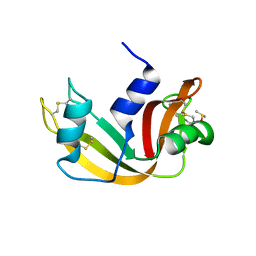 | |
4XLR
 
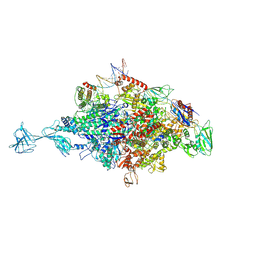 | |
1HLL
 
 | |
1HOF
 
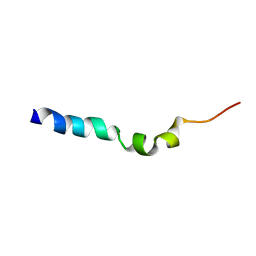 | |
6H4C
 
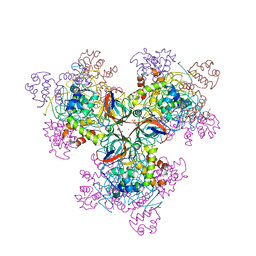 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
6H48
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20 | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
6H49
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
