3EUH
 
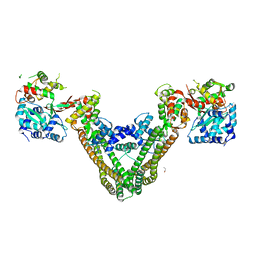 | | Crystal Structure of the MukE-MukF Complex | | Descriptor: | Chromosome partition protein mukF, GLYCINE, MukE | | Authors: | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
4IXU
 
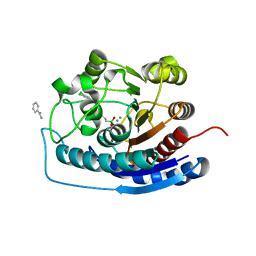 | | Crystal structure of human Arginase-2 complexed with inhibitor 11d: {(5R)-5-amino-5-carboxy-5-[(3-endo)-8-(3,4-dichlorobenzyl)-8-azabicyclo[3.2.1]oct-3-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7BJW
 
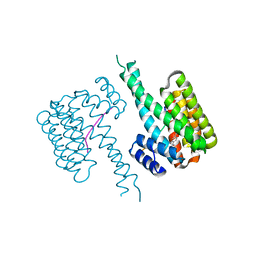 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-154 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-(4-oxidanylpiperidin-1-yl)benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJF
 
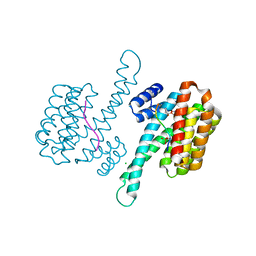 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-069 | | Descriptor: | (5-methyl-2-nitro-phenyl) cyclobutanecarboxylate, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
1V3H
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
3THZ
 
 | | Human MutSbeta complexed with an IDL of 6 bases (Loop6) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop6 minus strand, DNA Loop6 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Mechanism of mismatch repair revealed by human MutS bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
7BIQ
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-180 | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{R})-2-methyl-3,4-dihydro-2~{H}-quinolin-1-yl]sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
4IZV
 
 | |
1BHE
 
 | |
1BA1
 
 | |
7BKH
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-153 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-[(3S)-3-oxidanylpiperidin-1-yl]benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
1B3V
 
 | | XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOSE | | Descriptor: | PROTEIN (XYLANASE), alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Schmidt, A, Kratky, C. | | Deposit date: | 1998-12-15 | | Release date: | 1999-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant.
Biochemistry, 38, 1999
|
|
6D6X
 
 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
4J0N
 
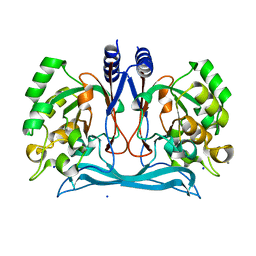 | | Crystal structure of a manganese dependent isatin hydrolase | | Descriptor: | CALCIUM ION, Isatin hydrolase B, MANGANESE (II) ION, ... | | Authors: | Bjerregaard-Andersen, K, Sommer, T, Jensen, J.K, Jochimsen, B, Etzerodt, M, Morth, J.P. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A proton wire and water channel revealed in the crystal structure of isatin hydrolase.
J.Biol.Chem., 289, 2014
|
|
1AP8
 
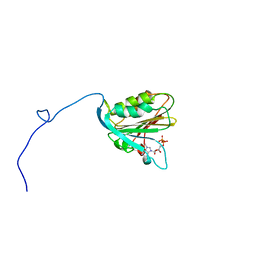 | | TRANSLATION INITIATION FACTOR EIF4E IN COMPLEX WITH M7GDP, NMR, 20 STRUCTURES | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR EIF4E | | Authors: | Matsuo, H, Li, H, Mcguire, A.M, Fletcher, M, Gingras, A.C, Sonenberg, N, Wagner, G. | | Deposit date: | 1997-07-25 | | Release date: | 1998-01-28 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein.
Nat.Struct.Biol., 4, 1997
|
|
4J1A
 
 | |
7BI3
 
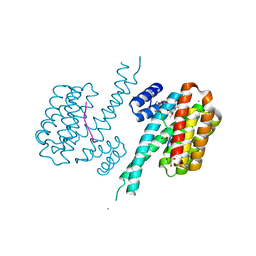 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-179 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
3OIB
 
 | |
3TE7
 
 | | Quinone Oxidoreductase (NQ02) bound to the imidazoacridin-6-one 5a1 | | Descriptor: | 5-{[2-(dimethylamino)ethyl]amino}-8-methoxy-6H-imidazo[4,5,1-de]acridin-6-one, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Dunstan, M.S. | | Deposit date: | 2011-08-12 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Inhibitors of NRH:Quinone Oxidoreductase 2 (NQO2): Crystal Structures, Biochemical Activity, and Intracellular Effects of Imidazoacridin-6-ones.
J.Med.Chem., 54, 2011
|
|
3OVP
 
 | | Crystal Structure of hRPE | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, FE (II) ION, Ribulose-phosphate 3-epimerase | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
1O6K
 
 | | Structure of activated form of PKB kinase domain S474D with GSK3 peptide and AMP-PNP | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
7BIY
 
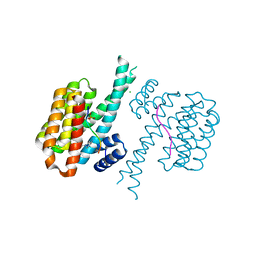 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-175 | | Descriptor: | 1-(4-methanoylphenyl)carbonylpiperidine-4-carbonitrile, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BA7
 
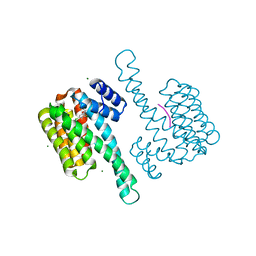 | | Cys-42-tethered stabilizer 9 of 14-3-3(sigma)/ERa PPI | | Descriptor: | 14-3-3 protein sigma, 2-[3,5-bis(chloranyl)phenoxy]-2-methyl-~{N}-(2-sulfanylethyl)propanamide, Estrogen receptor, ... | | Authors: | Sijbesma, E, Ottmann, C. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of a 14-3-3 PPI Pocket by Covalent Fragments as Stabilizers.
Acs Med.Chem.Lett., 12, 2021
|
|
7BA5
 
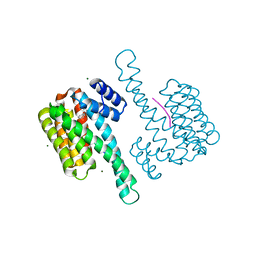 | | Cys-42-tethered stabilizer 7 of 14-3-3(sigma)/ERa PPI | | Descriptor: | 14-3-3 protein sigma, 2-(4-fluoranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, Estrogen receptor, ... | | Authors: | Sijbesma, E, Ottmann, C. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of a 14-3-3 PPI Pocket by Covalent Fragments as Stabilizers.
Acs Med.Chem.Lett., 12, 2021
|
|
3OLD
 
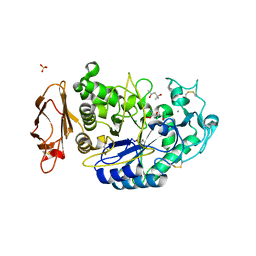 | | Crystal structure of alpha-amylase in complex with acarviostatin I03 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Qin, X, Ren, L. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes
J.Struct.Biol., 174, 2011
|
|
