1Z84
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana at5g18200 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ZINC ION, ... | | Authors: | Mccoy, J.G, Bitto, E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-03-29 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
3RWQ
 
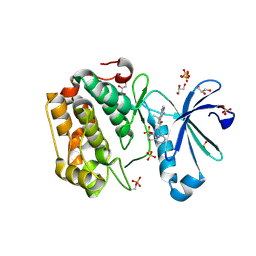 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Kazmirski, S, Kohls, D. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
1BHA
 
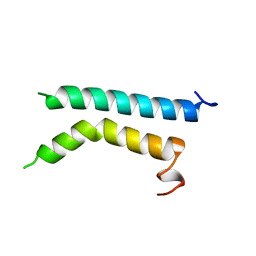 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
1FHJ
 
 | |
1FI1
 
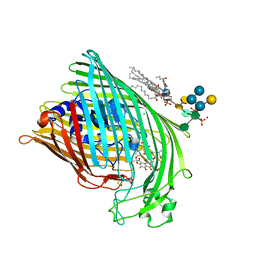 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
2MPF
 
 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1ZKK
 
 | | Crystal structure of hSET8 in ternary complex with H4 peptide (16-24) and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase, H4 lysine-20 specific, Peptide corresponding to residues 15-24 of histone H4, ... | | Authors: | Couture, J.-F, Collazo, E, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
Genes Dev., 19, 2005
|
|
1FGR
 
 | |
1Q8G
 
 | | NMR structure of human Cofilin | | Descriptor: | Cofilin, non-muscle isoform | | Authors: | Pope, B.J, Zierler-Gould, K.M, Kuhne, R, Weeds, A.G, Ball, L.J. | | Deposit date: | 2003-08-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human cofilin: rationalizing actin binding and pH sensitivity
J.Biol.Chem., 279, 2004
|
|
1Z0N
 
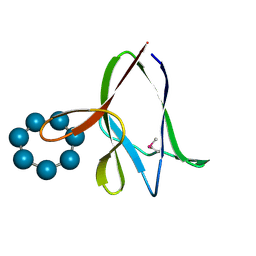 | | the glycogen-binding domain of the AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
1BW0
 
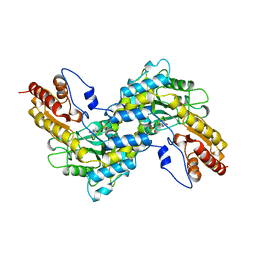 | | CRYSTAL STRUCTURE OF TYROSINE AMINOTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | PROTEIN (TYROSINE AMINOTRANSFERASE) | | Authors: | Blankenfeldt, W, Montemartini, M, Hunter, G.R, Kalisz, H.M, Nowicki, C, Hecht, H.J. | | Deposit date: | 1998-09-28 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode.
Protein Sci., 8, 1999
|
|
1Z88
 
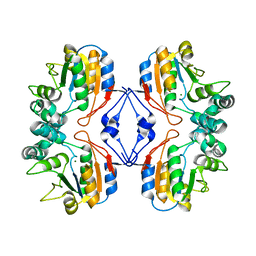 | |
1BT4
 
 | |
3SBK
 
 | | Russell's viper venom serine proteinase, RVV-V (PPACK-bound form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Vipera russelli proteinase RVV-V gamma | | Authors: | Nakayama, D, Ben Ammar, Y, Takeda, S. | | Deposit date: | 2011-06-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of coagulation factor V recognition for cleavage by RVV-V
Febs Lett., 585, 2011
|
|
2N4J
 
 | |
1FBT
 
 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | Authors: | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
1BT5
 
 | | CRYSTAL STRUCTURE OF THE IMIPENEM INHIBITED TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Maveyraud, L, Mourey, L, Pedelacq, J.D, Guillet, V, Kotra, L.K, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A Beta-Lactamases from Antibiotic-Resistant Bacteria
J.Am.Chem.Soc., 120, 1998
|
|
2MZ8
 
 | | Solution NMR structure of Salmonella Typhimurium transcriptional regulator protein Crl | | Descriptor: | Sigma factor-binding protein Crl | | Authors: | Cavaliere, P, Levi-Acobas, F, Monteil, V, Bellalou, J, Mayer, C, Norel, F, Sizun, C. | | Deposit date: | 2015-02-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding interface between the Salmonella sigma (S)/RpoS subunit of RNA polymerase and Crl: hints from bacterial species lacking crl.
Sci Rep, 5, 2015
|
|
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1BXV
 
 | | REDUCED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
1QBV
 
 | | CRYSTAL STRUCTURE OF THROMBIN COMPLEXED WITH AN GUANIDINE-MIMETIC INHIBITOR | | Descriptor: | Hirudin, THROMBIN (HEAVY CHAIN), THROMBIN (LIGHT CHAIN), ... | | Authors: | Bone, R, Lu, T, Illig, C.R, Soll, R.M, Spurlino, J.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-10-27 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of thrombin complexed with potent inhibitors incorporating a phenyl group as a peptide mimetic and aminopyridines as guanidine substitutes.
J.Med.Chem., 41, 1998
|
|
1C1Z
 
 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | Authors: | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
3SRE
 
 | | Serum paraoxonase-1 by directed evolution at pH 6.5 | | Descriptor: | BROMIDE ION, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ben David, M, Elias, M, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.
J.Mol.Biol., 418, 2012
|
|
2N8R
 
 | |
1UGF
 
 | |
