1VPE
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF PHOSPHOGLYCERATE KINASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Auerbach, G, Huber, R, Graettinger, M, Zaiss, K, Schurig, H, Jaenicke, R, Jacob, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.
Structure, 5, 1997
|
|
1VPF
 
 | | STRUCTURE OF HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR | | Authors: | Muller, Y.A, De Vos, A.M. | | Deposit date: | 1997-04-08 | | Release date: | 1998-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1VPH
 
 | |
1VPI
 
 | | PHOSPHOLIPASE A2 INHIBITOR FROM VIPOXIN | | Descriptor: | PHOSPHOLIPASE A2 INHIBITOR | | Authors: | Devedjiev, Y.D, Popov, A.N. | | Deposit date: | 1996-12-17 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray structure at 1.76 A resolution of a polypeptide phospholipase A2 inhibitor.
J.Mol.Biol., 266, 1997
|
|
1VPK
 
 | |
1VPL
 
 | |
1VPM
 
 | |
1VPN
 
 | | UNASSEMBLED POLYOMAVIRUS VP1 PENTAMER | | Descriptor: | POLYOMAVIRUS VP1 PENTAMER | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding.
Embo J., 16, 1997
|
|
1VPO
 
 | | Crystal Structure Analysis of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | TESTOSTERONE, anti-testosterone (heavy chain), anti-testosterone (light chain) | | Authors: | Valjakka, J.M, Hemminki, A, Niemi, S, Soderlund, H, Takkinen, K, Rouvinen, J. | | Deposit date: | 2004-11-15 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of an in Vitro Affinity- and Specificity-matured Anti-testosterone
Fab in Complex with Testosterone. IMPROVED AFFINITY RESULTS FROM
SMALL STRUCTURAL CHANGES WITHIN THE VARIABLE DOMAINS
J.Biol.Chem., 277, 2002
|
|
1VPP
 
 | | COMPLEX BETWEEN VEGF AND A RECEPTOR BLOCKING PEPTIDE | | Descriptor: | PROTEIN (PEPTIDE V108), PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR) | | Authors: | Wiesmann, C, Christinger, H.W, Cochran, A.G, Cunningham, B.C, Fairbrother, W.J, Keenan, C.J, Meng, G, de Vos, A.M. | | Deposit date: | 1998-10-09 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex between VEGF and a receptor-blocking peptide.
Biochemistry, 37, 1998
|
|
1VPQ
 
 | |
1VPR
 
 | |
1VPS
 
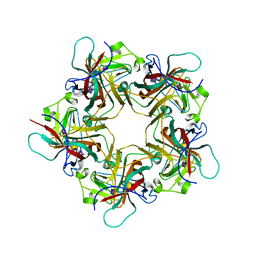 | | POLYOMAVIRUS VP1 PENTAMER COMPLEXED WITH A DISIALYLATED HEXASACCHARIDE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, POLYOMAVIRUS VP1 PENTAMER | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding.
Embo J., 16, 1997
|
|
1VPT
 
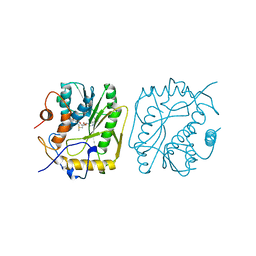 | | AS11 VARIANT OF VACCINIA VIRUS PROTEIN VP39 IN COMPLEX WITH S-ADENOSYL-L-METHIONINE | | Descriptor: | S-ADENOSYLMETHIONINE, VP39 | | Authors: | Hodel, A.E, Gershon, P.D, Shi, X, Quiocho, F.A. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VPU
 
 | |
1VPV
 
 | |
1VPW
 
 | | STRUCTURE OF THE PURR MUTANT, L54M, BOUND TO HYPOXANTHINE AND PURF OPERATOR DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PURINE REPRESSOR | | Authors: | Arvidson, D.N, Lu, F, Faber, C, Zalkin, H, Brennan, R.G. | | Deposit date: | 1998-02-27 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of PurR mutant L54M shows an alternative route to DNA kinking.
Nat.Struct.Biol., 5, 1998
|
|
1VPX
 
 | |
1VPY
 
 | |
1VPZ
 
 | |
1VQ0
 
 | |
1VQ1
 
 | |
1VQ2
 
 | | CRYSTAL STRUCTURE OF T4-BACTERIOPHAGE DEOXYCYTIDYLATE DEAMINASE, MUTANT R115E | | Descriptor: | 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, DEOXYCYTIDYLATE DEAMINASE, ZINC ION | | Authors: | Almog, R, Maley, F, Maley, G.F, Maccoll, R, Van Roey, P. | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of the R115E Mutant of T4-Bacteriophage 2'-Deoxycytidylate Deaminase
Biochemistry, 43, 2004
|
|
1VQ3
 
 | |
1VQ4
 
 | |
