6G2E
 
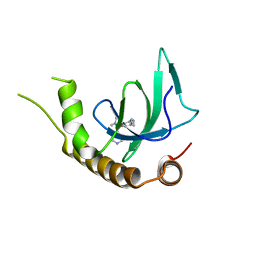 | | X-ray structure of NSD3-PWWP1 in complex with compound 13 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
5HLK
 
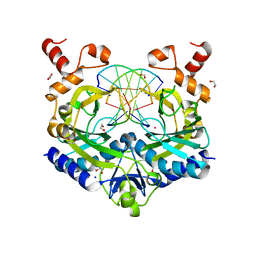 | | Crystal structure of the ternary EcoRV-DNA-Lu complex with cleaved DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*AP*GP*AP*TP)-3'), DNA (5'-D(*AP*TP*CP*TP*TP*TP)-3'), ... | | Authors: | Sangani, S.S, Kehr, A.D, Sinha, K, Rule, G.S, Jen-Jacobson, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Ion Binding at the Catalytic Site Induces Widely Distributed Changes in a Sequence Specific Protein-DNA Complex.
Biochemistry, 55, 2016
|
|
6RSC
 
 | |
5HX5
 
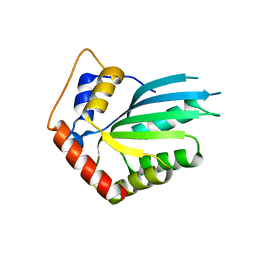 | | APOBEC3F Catalytic Domain Crystal Structure | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | Authors: | Shaban, N.M, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 1.92 Angstrom Zinc-Free APOBEC3F Catalytic Domain Crystal Structure.
J.Mol.Biol., 428, 2016
|
|
6A3J
 
 | | Levoglucosan dehydrogenase, complex with NADH and L-sorbose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative dehydrogenase, ... | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6VLB
 
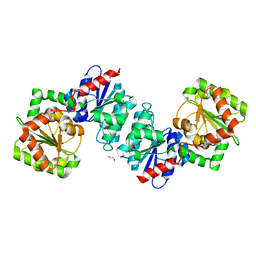 | |
4EMV
 
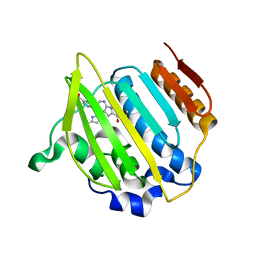 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 5-{2-(ethylcarbamoyl)-4-[3-(trifluoromethyl)-1H-pyrazol-1-yl]-1H-pyrrolo[2,3-b]pyridin-5-yl}pyridine-3-carboxylic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J, Hull, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6S17
 
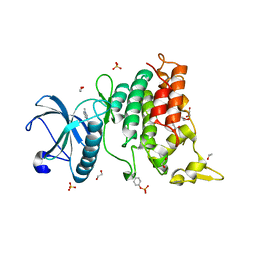 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
5HXA
 
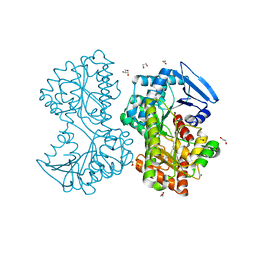 | |
6GBL
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
5HNN
 
 | |
6S5U
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with N-[2-(1H-Indol-3-yl)ethyl]-3-methyl-1-butanamine | | Descriptor: | Strictosidine synthase, ~{N}-[2-(1~{H}-indol-3-yl)ethyl]-3-methyl-butan-1-amine | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S7Z
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-((3,4-Dihydroisoquinolin-2(1H)-yl)sulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S1J
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(3~{S})-3-fluoranylpyrrolidin-1-yl]pyrimidin-4-yl]pyrazolo[1,5-b]pyridazine, DIMETHYL SULFOXIDE, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
7WH1
 
 | | structure of C elegans BCMO-2 | | Descriptor: | Beta-Carotene 15,15'-MonoOxygenase, FE (III) ION, GLYCEROL, ... | | Authors: | Pan, W, Liu, L. | | Deposit date: | 2021-12-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Nonheme Iron Enzymes BCMO-1 and BCMO-2 from Caenorhabditis elegans .
Front Mol Biosci, 9, 2022
|
|
5EJR
 
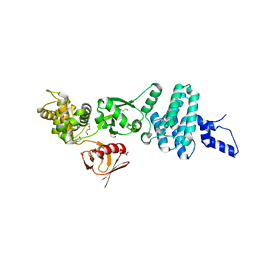 | | Structure of Dictyostelium Discoideum Myosin VII MyTH4-FERM MF2 domain | | Descriptor: | 1,2-ETHANEDIOL, Myosin-I heavy chain | | Authors: | Planelles-Herrero, V.J, Sirkia, H, Sourigues, Y, Clause, J, Titus, M.A, Houdusse, A. | | Deposit date: | 2015-11-02 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Myosin MyTH4-FERM structures highlight important principles of convergent evolution.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7WAM
 
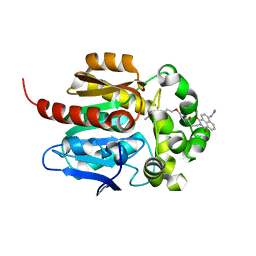 | | Crystal structure of HaloTag complexed with VL1 | | Descriptor: | 3-[6-(2-azanylhydrazinyl)-1,3-bis(oxidanylidene)benzo[de]isoquinolin-2-yl]-N-[2-(2-hexoxyethoxy)ethyl]propanamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Pratyush, M, Kang, M, Lee, H, Lee, C, Rhee, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A chemical tool for blue light-inducible proximity photo-crosslinking in live cells.
Chem Sci, 13, 2022
|
|
5HX4
 
 | |
6CE6
 
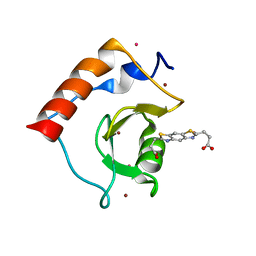 | | Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid | | Descriptor: | 3,3'-(benzo[1,2-d:5,4-d']bis[1,3]thiazole-2,6-diyl)dipropanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
7D7S
 
 | |
7FWR
 
 | | Crystal Structure of human FABP4 in complex with 3-[(3,4-dichlorophenyl)methylsulfanyl]-1,2,4-triazin-5-ol, i.e. SMILES c1(CSc2nc(cnn2)O)cc(c(cc1)Cl)Cl with IC50=0.290 microM | | Descriptor: | 3-{[(3,4-dichlorophenyl)methyl]sulfanyl}-1,2,4-triazin-5-ol, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Montavon, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FXY
 
 | | Crystal Structure of human FABP4 in complex with (4-chlorophenyl)methyl 5-fluoro-2,4-dioxo-1H-pyrimidine-6-carboxylate, i.e. SMILES C1(=C(C(=O)NC(=O)N1)F)C(=O)OCc1ccc(cc1)Cl with IC50=1.9 microM | | Descriptor: | (4-chlorophenyl)methyl 5-fluoro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylate, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kiss, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FWO
 
 | | Crystal Structure of human FABP4 in complex with 2-cyclohexyl-6-hydroxy-1-benzofuran-3-one | | Descriptor: | (2S)-2-cyclohexyl-6-hydroxy-1-benzofuran-3(2H)-one, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Boehringer, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FXT
 
 | | Crystal Structure of human FABP4 in complex with N-(2,1,3-benzothiadiazol-4-yl)-2,5-dichlorothiophene-3-sulfonamide, i.e. SMILES c12=NSN=c2cccc1NS(=O)(=O)C1=C(SC(=C1)Cl)Cl with IC50=0.255 microM | | Descriptor: | FORMIC ACID, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FZ6
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2S)-2-(phenoxymethyl)cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@H]([C@H](C1)C(=O)O)COc1ccccc1 with IC50=5.74145 microM | | Descriptor: | (1S,2R)-2-(phenoxymethyl)cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
