3NPN
 
 | |
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
2QH4
 
 | |
2QH3
 
 | |
2QH2
 
 | |
2M17
 
 | |
2M1V
 
 | |
2KTQ
 
 | | OPEN TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Li, Y, Waksman, G. | | Deposit date: | 1998-07-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation.
EMBO J., 17, 1998
|
|
2R8G
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DT)-3', 5'-D(*DTP*DCP*DAP*DCP*(P)P*DGP*DAP*DAP*DAP*DTP*DCP*DCP*DTP*DTP*DCP*DCP*DCP*DCP*DC)-3', ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
2R8I
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
2R8H
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
2JTP
 
 | |
3HYF
 
 | | Crystal structure of HIV-1 RNase H p15 with engineered E. coli loop and active site inhibitor | | Descriptor: | 2-(3,4-dichlorobenzyl)-5,6-dihydroxypyrimidine-4-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information.
J.Med.Chem., 52, 2009
|
|
8Q3R
 
 | | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component OPG148, Uracil-DNA glycosylase | | Authors: | Burmeister, W.P, Ballandras-Colas, A, Boettcher, B, Grimm, C. | | Deposit date: | 2023-08-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Plos Pathog., 20, 2024
|
|
2NCI
 
 | |
8TDZ
 
 | |
8TE0
 
 | |
8TDY
 
 | |
8TE2
 
 | |
8BS7
 
 | |
2PN9
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA modified aptamer | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*C)-3', RNA 16-mer with locked residues 9-10 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.-J. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
4OD8
 
 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
4ODA
 
 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
6ZXP
 
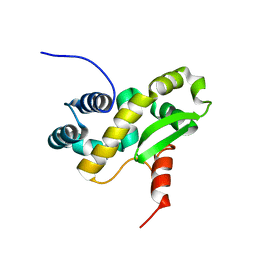 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20 fused to a short peptide from the viral DNA polymerase E9. | | Descriptor: | DNA polymerase processivity factor component A20,DNA polymerase processivity factor component E9 | | Authors: | Bersch, B, Tarbouriech, N, Burmeister, W, Iseni, F. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZYC
 
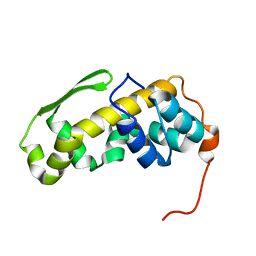 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20. | | Descriptor: | DNA polymerase processivity factor component A20 | | Authors: | Bersch, B, Iseni, F, Burmeister, W, Tarbouriech, N. | | Deposit date: | 2020-07-31 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
