5H4E
 
 | |
6IBY
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 6 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpyrrolidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7VB5
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum complexed with acetone cyanohydrin | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXY-2-METHYLPROPANENITRILE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
7VB3
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aliphatic (R)-hydroxynitrile lyase, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
6QQR
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 1~{H}-indol-5-ylboronic acid, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
5L72
 
 | |
6QMD
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{R})-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
7N3T
 
 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
9INF
 
 | | Cryo-EM structure of human XPR1 in closed state in the presence of KIDINS220-1-432 and 10 mM KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Yin, Y, Zuo, P, Liang, L. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
7EHT
 
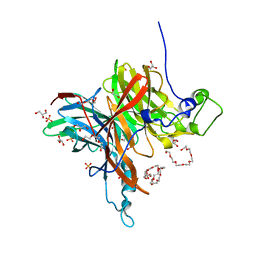 | | Levansucrase from Brenneria sp. EniD 312 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Levansucrase, ... | | Authors: | Xu, W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
8SGN
 
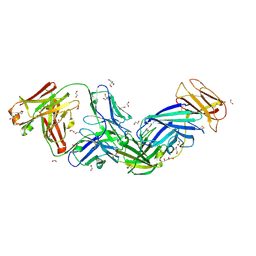 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
7EHR
 
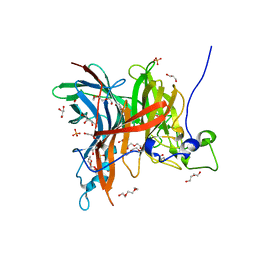 | | Levansucrase from Brenneria sp. EniD 312 at 1.33 angstroms resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Xu, W, Ni, D.W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
4ZXC
 
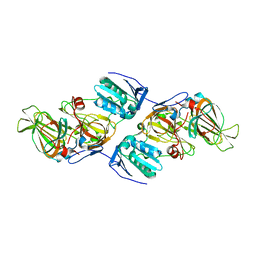 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Fe3+ | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
6QME
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chloranyl-3-methyl-phenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
9BFE
 
 | | Tyrocidine synthetase modules 1 and 2 crosslinked in the condensation state, complex B | | Descriptor: | (7S,10aR)-1-methyl-4-{4-[(5R)-1,1,5-trihydroxy-4,4-dimethyl-1,6,11-trioxo-2-oxa-7,10-diaza-1lambda~5~-phosphadodecan-12-yl]phenyl}-3,5,6,7,8,9,10,10a-octahydrocycloocta[d]pyridazin-7-yl [2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]carbamate, ADENOSINE MONOPHOSPHATE, Tyrocidine synthase 1, ... | | Authors: | Heberlig, G.W, Burkart, M.D. | | Deposit date: | 2024-04-17 | | Release date: | 2024-10-09 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Crosslinking intermodular condensation in non-ribosomal peptide biosynthesis.
Nature, 638, 2025
|
|
9BFD
 
 | | Tyrocidine synthetase modules 1 and 2 crosslinked in the condensation state, complex A | | Descriptor: | (7S,10aR)-1-methyl-4-{4-[(5R)-1,1,5-trihydroxy-4,4-dimethyl-1,6,11-trioxo-2-oxa-7,10-diaza-1lambda~5~-phosphadodecan-12-yl]phenyl}-3,5,6,7,8,9,10,10a-octahydrocycloocta[d]pyridazin-7-yl [2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]carbamate, 5'-O-[(R)-hydroxy{[(2S)-pyrrolidin-2-ylcarbonyl]oxy}phosphoryl]adenosine, Tyrocidine synthase 1, ... | | Authors: | Heberlig, G.W, Burkart, M.D. | | Deposit date: | 2024-04-17 | | Release date: | 2024-10-09 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Crosslinking intermodular condensation in non-ribosomal peptide biosynthesis.
Nature, 638, 2025
|
|
8XAM
 
 | | Co-crystal structure of compound 7 in complex with MAT2A | | Descriptor: | 2-[3-[7-chloranyl-4-(dimethylamino)-2-oxidanylidene-quinazolin-1-yl]phenoxy]-~{N}-[3-[7-chloranyl-4-(dimethylamino)-2-oxidanylidene-quinazolin-1-yl]phenyl]ethanamide, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Gao, F, Ding, X. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of novel MAT2A inhibitors by an allosteric site-compatible fragment growing approach.
Bioorg.Med.Chem., 100, 2024
|
|
5UGF
 
 | | Crystal structure of human purine nucleoside phosphorylase (F159Y) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Cameron, S.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-01-08 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic-site design for inverse heavy-enzyme isotope effects in human purine nucleoside phosphorylase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6IHH
 
 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
7BCD
 
 | | Notum Fragment 714 | | Descriptor: | 2-(morpholin-4-ium-4-ylmethyl)naphthalen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
9HNC
 
 | | Crystal structure of potassium-independent L-asparaginase from Phaseolus vulgaris (PvAIII, PvAspG2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Loch, J.I, Pierog, I, Imiolczyk, B, Barciszewski, J, Marsolais, F, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Unique double-helical packing of protein molecules in the crystal of potassium-independent L-asparaginase from common bean.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9ENQ
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
5ND6
 
 | | Crystal structure of apo transketolase from Chlamydomonas reinhardtii | | Descriptor: | 1,2-ETHANEDIOL, Transketolase | | Authors: | Fermani, S, Zaffagnini, M, Francia, F, Pasquini, M. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the magnesium-dependent activation of transketolase from Chlamydomonas reinhardtii.
Biochim. Biophys. Acta, 1861, 2017
|
|
7BC9
 
 | | Notum Fragment 690 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-N-(4-phenyl-1,3-thiazol-2-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-18 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
5KY0
 
 | | mouse POFUT1 in complex with mouse Notch1 EGF12(D464G) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
