4Z2Q
 
 | | The crystal structure of Sclerotium Rolfsii lectin variant 1 (SSR1) in complex with N-acetyl-glucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
6EFX
 
 | |
8Z6Q
 
 | |
3HO2
 
 | | Structure of E.coli FabF(C163A) in complex with Platencin | | Descriptor: | 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-06-01 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8TNV
 
 | | Hemocyanin Functional Unit CCHB-g of Concholepas concholepas | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Munoz, S, Vallejos-Baccelliere, G, Manubens, A, Salazar, M, Nascimento, A.F.Z, Ambrosio, A.L.B, Becker, M.I, Guixe, V, Castro-Fernandez, V. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a functional unit from an immunogenic mollusk hemocyanin.
Structure, 32, 2024
|
|
6EIS
 
 | | DYRK1A in complex with JWC-055 | | Descriptor: | 1-[4-fluoranyl-2-(trifluoromethyl)phenyl]-9-(1~{H}-pyrazol-4-yl)benzo[h][1,6]naphthyridin-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3I2L
 
 | |
7BZJ
 
 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
7BF3
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
5JXE
 
 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
8R71
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HAdp4 collected at 1.22 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparinase, ZINC ION, ... | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6I46
 
 | | Structure of P. aeruginosa LpxC with compound 8: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-2-oxooxazol-3(2H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[5-(2-fluoranyl-4-methoxy-phenyl)-2-oxidanylidene-1,3-oxazol-3-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, GLYCEROL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
5MXW
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
6TPN
 
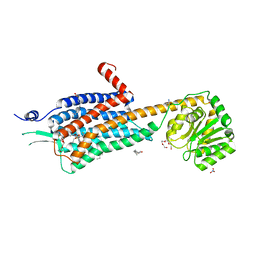 | | Crystal structure of the Orexin-2 receptor in complex with HTL6641 at 2.61 A resolution | | Descriptor: | 2-(5,6-dimethoxypyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, NITRATE ION, OLEIC ACID, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6Q9Y
 
 | |
5GXG
 
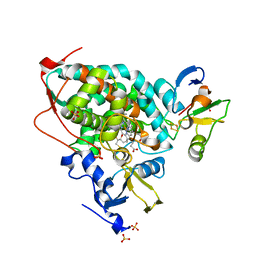 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8A1W
 
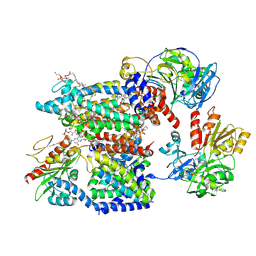 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q1 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
141D
 
 | |
8ZPA
 
 | | Crystal structure of a HSA/probe complex | | Descriptor: | 1,2-ETHANEDIOL, 7-$l^{3}-oxidanyl-12~{H}-[2,1]benzazaborolo[2,1-a][1,3,2]benzodiazaborinine, Albumin, ... | | Authors: | Youn, S.Y, Cha, S.S. | | Deposit date: | 2024-05-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Conversion of albumin into a BODIPY-like photosensitizer by a flick reaction, tumor accumulation and photodynamic therapy.
Biomaterials, 313, 2025
|
|
6E92
 
 | | CA IX mimic Complexed with Steroidal Sulfamate Compound STX 2845 | | Descriptor: | 6-(sulfamoyloxy)-2-[(3,4,5-trimethoxyphenyl)methyl]isoquinolin-2-ium, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, Mckenna, R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | 3,17 beta-Bis-sulfamoyloxy-2-methoxyestra-1,3,5(10)-triene and Nonsteroidal Sulfamate Derivatives Inhibit Carbonic Anhydrase IX: Structure-Activity Optimization for Isoform Selectivity.
J. Med. Chem., 62, 2019
|
|
8RLY
 
 | | E. coli endonuclease IV complexed with sulfate, catalytic Fe2+ | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Endonuclease 4, ... | | Authors: | Saper, M.A, Paterson, N.G, Kirillov, S, Rouvinski, A. | | Deposit date: | 2024-01-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli .
Biochemistry, 64, 2025
|
|
6TOK
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 23d | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1-methyl-3-(3-methyl-3-oxidanyl-butyl)benzimidazol-2-one, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5GG5
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hydrolase, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6BCB
 
 | |
