6LT8
 
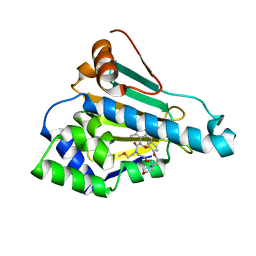 | | HSP90 in complex with KW-2478 | | Descriptor: | 2-[2-ethyl-6-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]carbonyl-3,5-bis(oxidanyl)phenyl]-~{N},~{N}-bis(2-methoxyethyl)ethanamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Anti-NSCLC activity in vitro of Hsp90 N inhibitor KW-2478 and complex crystal structure determination of Hsp90 N -KW-2478.
J.Struct.Biol., 213, 2021
|
|
6MAB
 
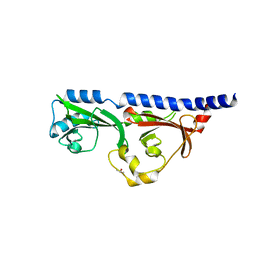 | | 1.7A resolution structure of RsbU from Chlamydia trachomatis (periplasmic domain) | | Descriptor: | ISOPROPYL ALCOHOL, Sigma regulatory family protein-PP2C phosphatase | | Authors: | Dmitriev, A, Lovell, S, Battaile, K.P, Soules, K, Hefty, P.S. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and ligand binding analyses of the periplasmic sensor domain of RsbU in Chlamydia trachomatis support a role in TCA cycle regulation.
Mol.Microbiol., 113, 2020
|
|
6MBB
 
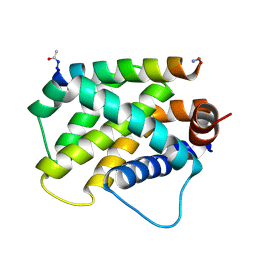 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
4UP1
 
 | |
8A6W
 
 | |
8A9P
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
3SB2
 
 | | Crystal Structure of the RNA chaperone Hfq from Herbaspirillum seropedicae SMR1 | | Descriptor: | GLYCEROL, Protein hfq | | Authors: | Kadowaki, M.A.S, Iulek, J, Barbosa, J.A.R.G, Pedrosa, F.O, Souza, E.M, Chubatsu, L.S, Monteiro, R.A, Steffens, M.B.R. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6301 Å) | | Cite: | Structural characterization of the RNA chaperone Hfq from the nitrogen-fixing bacterium Herbaspirillum seropedicae SmR1.
Biochim.Biophys.Acta, 1824, 2011
|
|
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8A8L
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment | | Descriptor: | 6-methoxy-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole, Cytochrome P450 protein, GLYCEROL, ... | | Authors: | Snee, M, Katariya, M, Levy, C. | | Deposit date: | 2022-06-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment
To Be Published
|
|
6TQQ
 
 | |
6TQP
 
 | | Structural insight into tanapoxvirus mediated inhibition of apoptosis | | Descriptor: | 16L protein, Bcl-2-binding component 3, isoforms 1/2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84940946 Å) | | Cite: | Structural insight into tanapoxvirus-mediated inhibition of apoptosis.
Febs J., 287, 2020
|
|
6TRR
 
 | |
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
6T76
 
 | | PII-like protein CutA from Nostoc sp. PCC 7120 in apo form | | Descriptor: | Periplasmic divalent cation tolerance protein, SULFATE ION | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
6T7E
 
 | | PII-like protein CutA from Nostoc sp. PCC7120 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Periplasmic divalent cation tolerance protein | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
7F25
 
 | |
7FEV
 
 | | Crystal structure of Old Yellow Enzyme6 (OYE6) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN binding | | Authors: | Singh, Y, Sharma, R, Mishra, M, Verma, P.K, Saxena, A.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Crystal structure of ArOYE6 reveals a novel C-terminal helical extension and mechanistic insights into the distinct class III OYEs from pathogenic fungi.
Febs J., 289, 2022
|
|
6RUQ
 
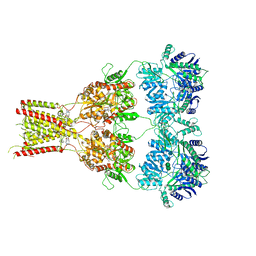 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
9NHF
 
 | |
6X3I
 
 | | NNAS Fc mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose | | Authors: | Wei, R, Zhou, Y.F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Engineered Fc-glycosylation switch to eliminate antibody effector function.
Mabs, 12, 2020
|
|
6HIL
 
 | |
8I5P
 
 | |
8I5Q
 
 | |
8I5T
 
 | |
8I5U
 
 | |
