1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
7ZJ1
 
 | | Crystal structure of ADAR1-dsRBD3 dimer | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Mboukou, A, Barraud, P. | | Deposit date: | 2022-04-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dimerization of ADAR1 modulates site-specificity of RNA editing
Biorxiv, 2023
|
|
1QBJ
 
 | | CRYSTAL STRUCTURE OF THE ZALPHA Z-DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), PROTEIN (DOUBLE-STRANDED RNA SPECIFIC ADENOSINE DEAMINASE (ADAR1)) | | Authors: | Schwartz, T, Rould, M.A, Rich, A. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA.
Science, 284, 1999
|
|
3F21
 
 | | Crystal structure of Zalpha in complex with d(CACGTG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DAP*DCP*DGP*DTP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
2GXB
 
 | | Crystal Structure of The Za Domain bound to Z-RNA | | Descriptor: | 5'-R(P*(DU)P*CP*GP*CP*GP*CP*G)-3', Double-stranded RNA-specific adenosine deaminase, SODIUM ION | | Authors: | Athanasiadis, A, Placido, D, Rich, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Left-Handed RNA Double Helix Bound by the Zalpha Domain of the RNA-Editing Enzyme ADAR1.
Structure, 15, 2007
|
|
7C0I
 
 | | Crystal structure of chimeric mutant of E3L in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-binding protein,Double-stranded RNA-specific adenosine deaminase, SULFATE ION | | Authors: | Choi, H.J, Park, C.H, Kim, J.S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
3F22
 
 | | Crystal structure of Zalpha in complex with d(CGTACG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DTP*DAP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
2ACJ
 
 | | Crystal structure of the B/Z junction containing DNA bound to Z-DNA binding proteins | | Descriptor: | 5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*GP*GP*CP*GP*CP*GP*CP*G)-3', 5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*AP*TP*AP*AP*AP*CP*C)-3', Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Lowenhaupt, K, Rich, A, Kim, Y.-G, Kim, K.K. | | Deposit date: | 2005-07-19 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases.
Nature, 437, 2005
|
|
3IRR
 
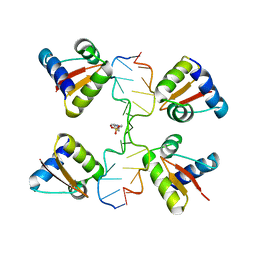 | | Crystal Structure of a Z-Z junction (with HEPES intercalating) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*A*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*G*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3F23
 
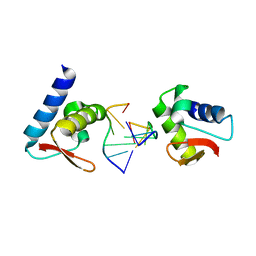 | | Crystal structure of Zalpha in complex with d(CGGCCG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DCP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
7C0J
 
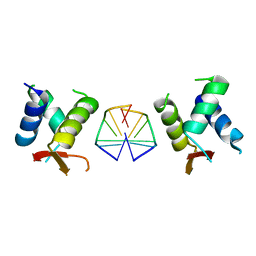 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7ZLQ
 
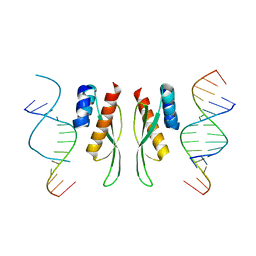 | |
3IRQ
 
 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ZUP
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*AP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZUO
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZU1
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
2L54
 
 | | Solution structure of the Zalpha domain mutant of ADAR1 (N43A,Y47A) | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Zhao, J, Pervushin, K, Feng, S, Droge, P. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Alternate rRNA secondary structures as regulators of translation
Nat.Struct.Mol.Biol., 18, 2011
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8GBD
 
 | | Homo sapiens Zalpha mutant - P193A | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Vogeli, B, Nichols, P.J, Henen, M, Vicens, Q. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8GBC
 
 | | Homo sapiens Zalpha mutant - N173S | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Nichols, P.J, Henen, M, Vicens, Q, Vogeli, B. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
2MDR
 
 | | Solution structure of the third double-stranded RNA-binding domain (dsRBD3) of human adenosine-deaminase ADAR1 | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Barraud, P, Banerjee, S, Mohamed, W.I, Jantsch, M.F, Allain, F.H. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
