5TC2
 
 | |
8TCJ
 
 | |
4Q1W
 
 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, ASPARTYL PROTEASE, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
5HRS
 
 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125A mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
6VQD
 
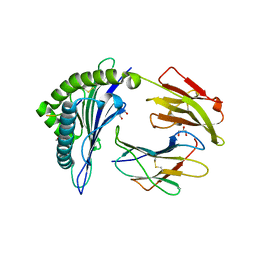 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
5HRR
 
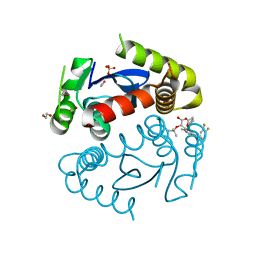 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125S mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
7XIS
 
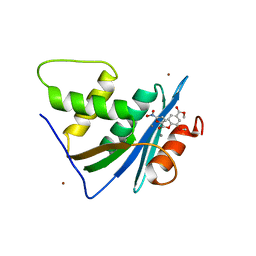 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
8F7M
 
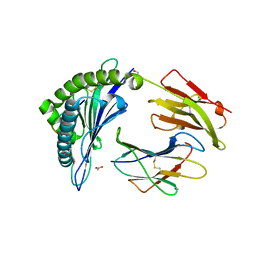 | |
7DT9
 
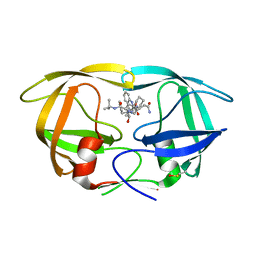 | | D30N HIV Protease in complex with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | D30N HIV Protease in complex with Saquinavir
To Be Published
|
|
6BJ3
 
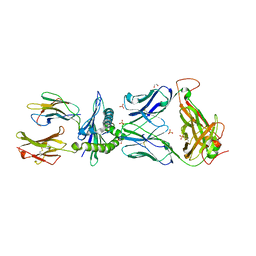 | | TCR55 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HIV Pol B35 peptide, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
4Q1X
 
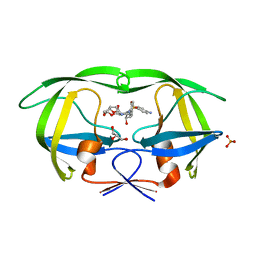 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ASPARTYL PROTEASE, GLYCEROL, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
1G6L
 
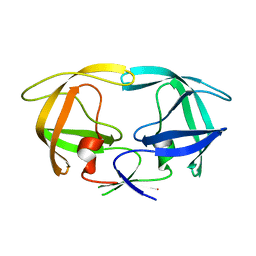 | |
1BWA
 
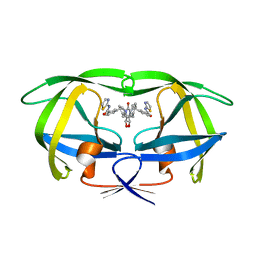 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
7DOZ
 
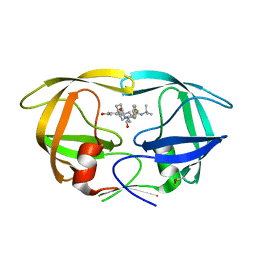 | | HIV-1 Protease D30N mutant in complex with Nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
J.Biomol.Struct.Dyn., 2021
|
|
8JYI
 
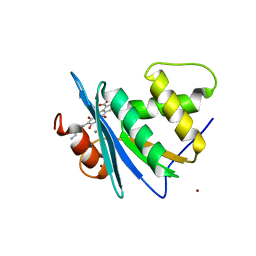 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
4U1I
 
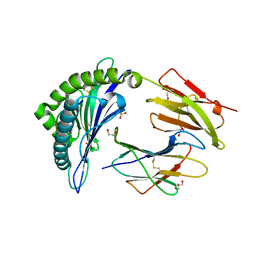 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
8F5A
 
 | |
8TCL
 
 | |
1ODY
 
 | | HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, HIV-1 PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | Authors: | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
1DMP
 
 | | STRUCTURE OF HIV-1 PROTEASE COMPLEX | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Chang, C.-H. | | Deposit date: | 1996-11-01 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450.
Chem.Biol., 3, 1996
|
|
3DOX
 
 | | X-ray structure of HIV-1 protease in situ product complex | | Descriptor: | A PEPTIDE SUBSTRATE-PIV, A PEPTIDE SUBSTRATE-SQNY, HIV-1 PROTEASE | | Authors: | Hosur, M.V, Ferrer, J.-L, Das, A, Prashar, V, Bihani, S. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of HIV-1 protease in situ product complex
Proteins, 74, 2009
|
|
1ODX
 
 | | HIV-1 Proteinase mutant A71T, V82A | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Kervinen, J, Thanki, N, Zdanov, A, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
6CPN
 
 | | Crystal structure of DR11 presenting the RQ13 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gag polyprotein, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
6J1V
 
 | | The structure of HLA-A*3003/RT313 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, RT313 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
