3BG7
 
 | |
6T8O
 
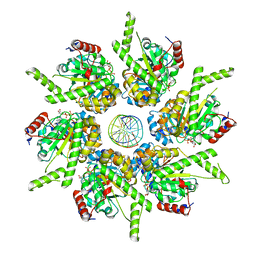 | | Stalled FtsK motor domain bound to dsDNA end | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, dsDNA substrate | | 著者 | Jean, N.L, Lowe, J. | | 登録日 | 2019-10-24 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3B96
 
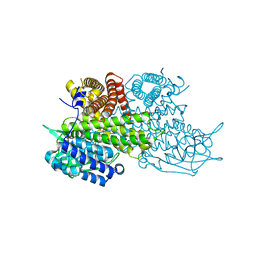 | | Structural Basis for Substrate Fatty-Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, TETRADECANOYL-COA, Very long-chain specific acyl-CoA dehydrogenase | | 著者 | McAndrew, R.P, Wang, Y, Mohsen, A.W, He, M, Vockley, J, Kim, J.J. | | 登録日 | 2007-11-02 | | 公開日 | 2008-02-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3B9O
 
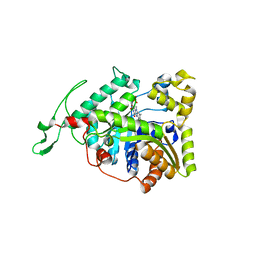 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | 分子名称: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | 著者 | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | 登録日 | 2007-11-06 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
1N52
 
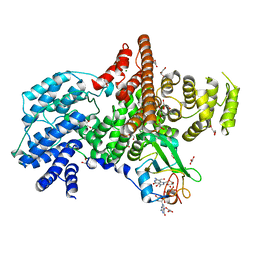 | | Cap Binding Complex | | 分子名称: | 20 kDa nuclear cap binding protein, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 80 kDa nuclear cap binding protein, ... | | 著者 | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | 登録日 | 2002-11-04 | | 公開日 | 2003-02-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
3T36
 
 | |
1N54
 
 | | Cap Binding Complex m7GpppG free | | 分子名称: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | 著者 | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | 登録日 | 2002-11-04 | | 公開日 | 2003-02-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
7REQ
 
 | |
6SNC
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LNO1 Heavy Chain, ... | | 著者 | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | 登録日 | 2019-08-23 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
1N5A
 
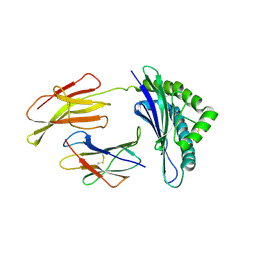 | | Crystal structure of the Murine class I Major Histocompatibility Complex of H-2DB, B2-Microglobulin, and A 9-Residue immunodominant peptide epitope gp33 derived from LCMV | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | 著者 | Achour, A, Michaelsson, J, Harris, R.A, Odeberg, J, Grufman, P, Sandberg, J.K, Levitsky, V, Karre, K, Sandalova, T, Schneider, G. | | 登録日 | 2002-11-05 | | 公開日 | 2003-01-07 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of
gp33 Revealed by Comparative Crystal Structure Analyses.
Immunity, 17, 2002
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | 分子名称: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | 著者 | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2011-05-30 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
6SKK
 
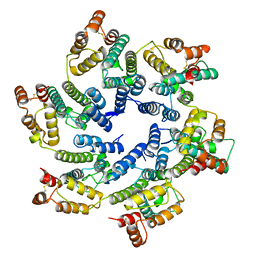 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | capsid protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3T6U
 
 | |
3T8I
 
 | | Structural analysis of thermostable S. solfataricus purine-specific nucleoside hydrolase | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minici, C, Cacciapuoti, G, De Leo, E, Porcelli, M, Degano, M. | | 登録日 | 2011-08-01 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | New Determinants in the Catalytic Mechanism of Nucleoside Hydrolases from the Structures of Two Isozymes from Sulfolobus solfataricus.
Biochemistry, 51, 2012
|
|
6BYK
 
 | |
6BX4
 
 | |
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | 分子名称: | Alkane monooxygenase | | 著者 | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | 登録日 | 2007-11-06 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3BAW
 
 | |
3Q14
 
 | | Toluene 4 monooxygenase HD Complex with p-cresol | | 分子名称: | FE (III) ION, P-CRESOL, PENTAETHYLENE GLYCOL, ... | | 著者 | Acheson, J.F, Bailey, L.J, Fox, B.G. | | 登録日 | 2010-12-16 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
6BZD
 
 | |
3BF8
 
 | |
3PI7
 
 | |
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | 分子名称: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2011-05-30 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
7RNI
 
 | |
3PPM
 
 | | Crystal Structure of a Noncovalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | 1-DODECANOL, 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptan-1-one, CHLORIDE ION, ... | | 著者 | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | 登録日 | 2010-11-24 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
