3AV5
 
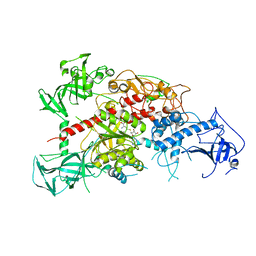 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | 著者 | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | 登録日 | 2011-02-22 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2A3V
 
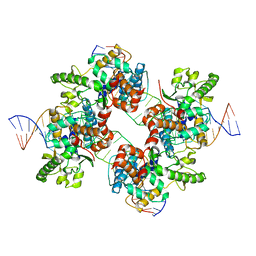 | | Structural basis for broad DNA-specificity in integron recombination | | 分子名称: | DNA (31-MER), DNA (34-MER), site-specific recombinase IntI4 | | 著者 | MacDonald, D, Demarre, G, Bouvier, M, Mazel, D, Gopaul, D.N. | | 登録日 | 2005-06-27 | | 公開日 | 2006-05-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for broad DNA-specificity in integron recombination.
Nature, 440, 2006
|
|
2JPA
 
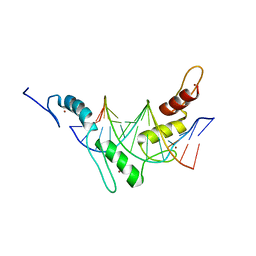 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | 分子名称: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | 著者 | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | 登録日 | 2007-05-01 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2LEX
 
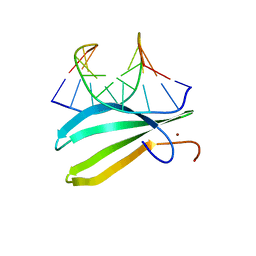 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | 分子名称: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | 著者 | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-06-24 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
6RYI
 
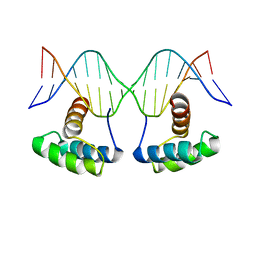 | | WUS-HD bound to G-Box DNA | | 分子名称: | DNA (5'-D(P*CP*CP*CP*AP*TP*CP*AP*CP*GP*TP*GP*AP*CP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*TP*CP*AP*CP*GP*TP*GP*AP*TP*GP*GP*G)-3'), Protein WUSCHEL | | 著者 | Sloan, J.J, Wild, K, Sinning, I. | | 登録日 | 2019-06-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.691 Å) | | 主引用文献 | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6S85
 
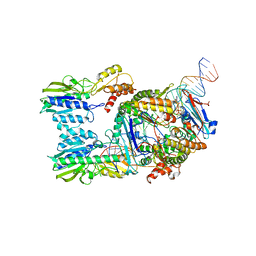 | | Cutting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), DNA (32-MER), ... | | 著者 | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | 登録日 | 2019-07-08 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
148D
 
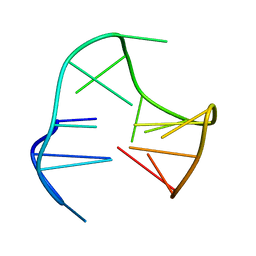 | |
177D
 
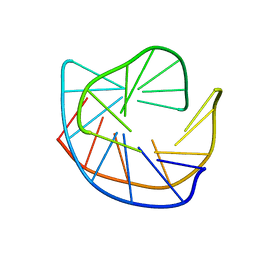 | |
2XNK
 
 | |
1AXU
 
 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | 分子名称: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | 著者 | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | 登録日 | 1997-10-21 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
4K74
 
 | |
3UQZ
 
 | |
6GZ7
 
 | | Polyamide - DNA complex NMR structure | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*G)-3'), dimethyl-[3-[3-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-propan-2-ylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoylamino]imidazol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]propyl]azanium | | 著者 | Aman, K. | | 登録日 | 2018-07-03 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Kinetic Profiling of Allosteric Modulation of Duplex DNA Induced by DNA-Binding Polyamide Analogues.
Chemistry, 25, 2019
|
|
7OHE
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*TP*CP*GP*AP*CP*GP*CP*G)-3') | | 著者 | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | 登録日 | 2021-05-10 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHM
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | 分子名称: | DNA (5'-D(*CP*GP*AP*(DH)P*GP*TP*CP*G)-3') | | 著者 | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | 登録日 | 2021-05-11 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHJ
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*CP*G)-3') | | 著者 | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | 登録日 | 2021-05-11 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OGV
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | 分子名称: | DNA (5'-D(*(P*GP*CP*GP*TP*(DH)P*GP*AP*CP*GP*CP*G-3') | | 著者 | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | 登録日 | 2021-05-07 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
1D21
 
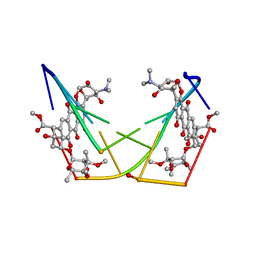 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | 分子名称: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), NOGALAMYCIN | | 著者 | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | 登録日 | 1990-08-08 | | 公開日 | 1991-07-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
8AV6
 
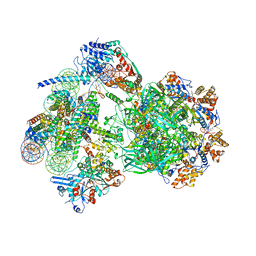 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-08-26 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
6RYD
 
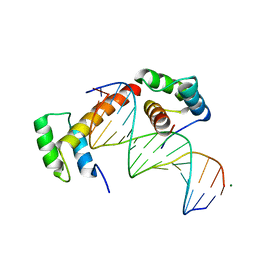 | | WUS-HD bound to TGAA DNA | | 分子名称: | DNA (5'-D(*AP*GP*TP*GP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*CP*AP*CP*T)-3'), MAGNESIUM ION, ... | | 著者 | Sloan, J.J, Wild, K, Sinning, I. | | 登録日 | 2019-06-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.575 Å) | | 主引用文献 | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RYL
 
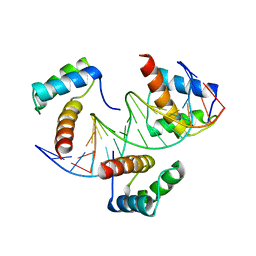 | | WUS-HD bound to TAAT DNA | | 分子名称: | DNA (5'-D(P*CP*AP*CP*AP*AP*CP*CP*CP*AP*TP*TP*AP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*TP*AP*AP*TP*GP*GP*GP*TP*TP*GP*TP*G)-3'), Protein WUSCHEL | | 著者 | Sloan, J.J, Wild, K, Sinning, I. | | 登録日 | 2019-06-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
8OR8
 
 | |
1D22
 
 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | 分子名称: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), U-58872, HYDROXY DERIVATIVE OF NOGALAMYCIN | | 著者 | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | 登録日 | 1990-08-08 | | 公開日 | 1991-07-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
2XNH
 
 | |
383D
 
 | |
