144L
 
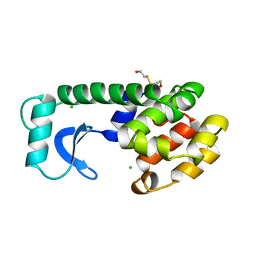 | |
1ANK
 
 | | THE CLOSED CONFORMATION OF A HIGHLY FLEXIBLE PROTEIN: THE STRUCTURE OF E. COLI ADENYLATE KINASE WITH BOUND AMP AND AMPPNP | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Berry, M.B, Meador, B, Bilderback, T, Liang, P, Glaser, M, Phillips Jr, G.N. | | 登録日 | 1994-02-28 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP.
Proteins, 19, 1994
|
|
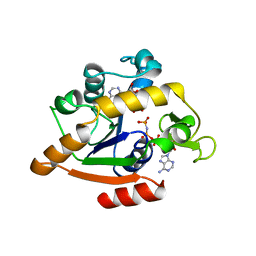
2WVD
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | 分子名称: | GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR, SULFATE ION | | 著者 | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | 登録日 | 2009-10-16 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
1F31
 
 | |
2X19
 
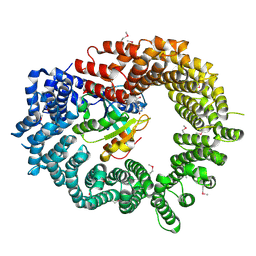 | | Crystal structure of Importin13 - RanGTP complex | | 分子名称: | GTP-BINDING NUCLEAR PROTEIN GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, IMPORTIN-13, ... | | 著者 | Bono, F, Cook, A.G, Gruenwald, M, Ebert, J, Conti, E. | | 登録日 | 2009-12-23 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nuclear Import Mechanism of the Ejc Component Mago- Y14 Revealed by Structural Studies of Importin 13.
Mol.Cell, 37, 2010
|
|
2WVE
 
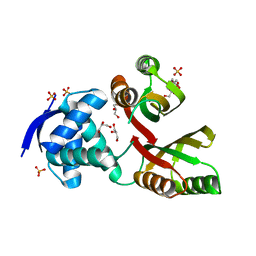 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, GLYCEROL, ... | | 著者 | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | 登録日 | 2009-10-16 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
3O35
 
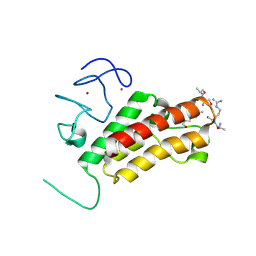 | |
1AB2
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY 2 DOMAIN OF C-ABL | | 分子名称: | C-ABL TYROSINE KINASE SH2 DOMAIN | | 著者 | Overduin, M, Rios, C.B, Mayer, B.J, Baltimore, D, Cowburn, D. | | 登録日 | 1993-07-19 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the src homology 2 domain of c-abl.
Cell(Cambridge,Mass.), 70, 1992
|
|
1BHG
 
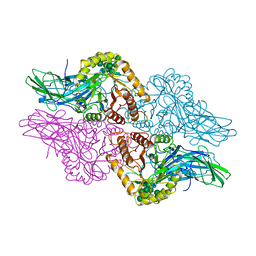 | | HUMAN BETA-GLUCURONIDASE AT 2.6 A RESOLUTION | | 分子名称: | BETA-GLUCURONIDASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Jain, S, Drendel, W.B. | | 登録日 | 1996-03-04 | | 公開日 | 1997-09-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs.
Nat.Struct.Biol., 3, 1996
|
|
1BEV
 
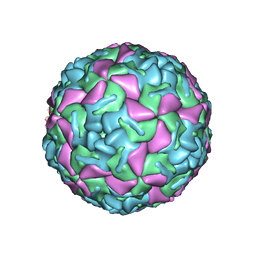 | | BOVINE ENTEROVIRUS VG-5-27 | | 分子名称: | BOVINE ENTEROVIRUS COAT PROTEINS VP1 TO VP4, MYRISTIC ACID, SULFATE ION | | 著者 | Smyth, M, Tate, J, Lyons, C, Hoey, E, Martin, S, Stuart, D. | | 登録日 | 1996-04-03 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Implications for viral uncoating from the structure of bovine enterovirus.
Nat.Struct.Biol., 2, 1995
|
|
1FHL
 
 | |
2KBB
 
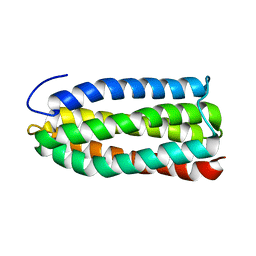 | | Solution Structure of the R9 Domain of Talin | | 分子名称: | Talin-1 | | 著者 | Goult, B.T, Gingras, A.R, Bate, N, Critchley, D.R, Barsukov, I.L. | | 登録日 | 2008-11-24 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of an interdomain complex that regulates talin activity.
J.Biol.Chem., 284, 2009
|
|
1BIT
 
 | |
2KGX
 
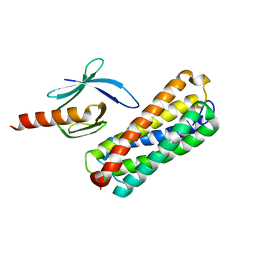 | | HADDOCK structure of the talin F3 domain in complex with talin 1655-1822 | | 分子名称: | MKIAA1027 protein, Talin-1 | | 著者 | Goult, B.T, Gingras, A.R, Bate, N, Critchley, D.R, Barsukov, I.L. | | 登録日 | 2009-03-23 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of an interdomain complex that regulates talin activity.
J.Biol.Chem., 284, 2009
|
|
1BEC
 
 | |
1F9K
 
 | | WINGED BEAN ACIDIC LECTIN COMPLEXED WITH METHYL-ALPHA-D-GALACTOSE | | 分子名称: | ACIDIC LECTIN, CALCIUM ION, MANGANESE (II) ION, ... | | 著者 | Manoj, N, Srinivas, V.R, Surolia, A, Vijayan, M, Suguna, K. | | 登録日 | 2000-07-11 | | 公開日 | 2001-07-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Carbohydrate specificity and salt-bridge mediated conformational change in acidic winged bean agglutinin.
J.Mol.Biol., 302, 2000
|
|
1BGS
 
 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | 分子名称: | BARNASE, BARSTAR | | 著者 | Guillet, V, Lapthorn, A, Mauguen, Y. | | 登録日 | 1993-11-02 | | 公開日 | 1994-04-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
3EYX
 
 | | Crystal structure of Carbonic Anhydrase Nce103 from Saccharomyces cerevisiae | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase, ... | | 著者 | Teng, Y.B, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | 登録日 | 2008-10-22 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural insights into the substrate tunnel of Saccharomyces cerevisiae carbonic anhydrase Nce103.
Bmc Struct.Biol., 9, 2009
|
|
1FKT
 
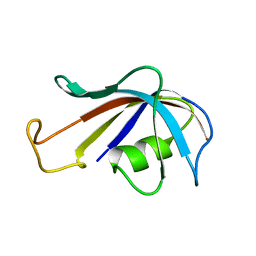 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | 分子名称: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | 著者 | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | 登録日 | 1992-03-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1BGT
 
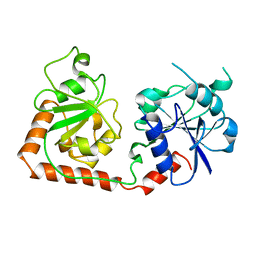 | |
1BGG
 
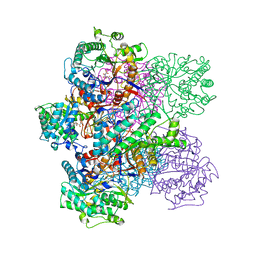 | | GLUCOSIDASE A FROM BACILLUS POLYMYXA COMPLEXED WITH GLUCONATE | | 分子名称: | BETA-GLUCOSIDASE A, D-gluconic acid | | 著者 | Sanz-Aparicio, J, Hermoso, J, Martinez-Ripoll, M, Polaina, J. | | 登録日 | 1997-05-12 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1AVY
 
 | | FIBRITIN DELETION MUTANT M (BACTERIOPHAGE T4) | | 分子名称: | FIBRITIN | | 著者 | Strelkov, S.V, Tao, Y, Mesyanzhinov, V.V, Rossmann, M.G. | | 登録日 | 1997-09-22 | | 公開日 | 1997-12-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of bacteriophage T4 fibritin M: a troublesome packing arrangement.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1FKR
 
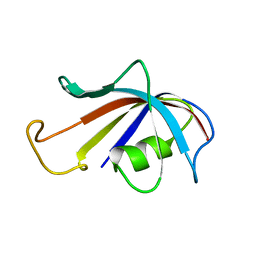 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | 分子名称: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | 著者 | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | 登録日 | 1992-03-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1QCS
 
 | |
1BK7
 
 | |
