6EMX
 
 | |
5J0S
 
 | |
5J0U
 
 | |
6HYX
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Y197F-P250C | | 分子名称: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel, ... | | 著者 | Hu, H.D, Delarue, M. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4BR6
 
 | | Crystal structure of Chaetomium thermophilum MnSOD | | 分子名称: | GLYCEROL, MANGANESE (III) ION, SODIUM ION, ... | | 著者 | Haikarainen, T, Frioux, C, Zhnag, L.-Q, Li, D.-C, Papageorgiou, A.C. | | 登録日 | 2013-06-04 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure and Biochemical Characterization of a Manganese Superoxide Dismutase from Chaetomium Thermophilum.
Biochim.Biophys.Acta, 1844, 2014
|
|
5OPD
 
 | |
5C1P
 
 | | Crystal structure of ADP and D-alanyl-D-alanine complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, ... | | 著者 | Tran, H.T, Kang, L.W, Hong, M.K, Ngo, H.P.T. | | 登録日 | 2015-06-15 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7CM1
 
 | | Neuraminidase from the Wuhan Asiatic toad influenza virus | | 分子名称: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | 著者 | Wang, J. | | 登録日 | 2020-07-23 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Lattice-translocation defects in specific crystals of the catalytic head domain of influenza neuraminidase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6EF9
 
 | | GspB Siglec domain | | 分子名称: | FORMIC ACID, Platelet binding protein GspB - Siglec domain, SODIUM ION | | 著者 | Iverson, T.M. | | 登録日 | 2018-08-16 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
4MR1
 
 | |
6B5E
 
 | | Mycobacterium tuberculosis RmlA in complex with dTDP-glucose | | 分子名称: | 1,2-ETHANEDIOL, 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CHLORIDE ION, ... | | 著者 | Brown, H.A, Holden, H.M. | | 登録日 | 2017-09-29 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The structure of glucose-1-phosphate thymidylyltransferase from Mycobacterium tuberculosis reveals the location of an essential magnesium ion in the RmlA-type enzymes.
Protein Sci., 27, 2018
|
|
4PMO
 
 | | Crystal structure of the Mycobacterium tuberculosis Tat-secreted protein Rv2525c, monoclinic crystal form I | | 分子名称: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | 著者 | Bellinzoni, M, Haouz, A, Shepard, W, Alzari, P.M. | | 登録日 | 2014-05-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
J.Struct.Biol., 188, 2014
|
|
5M67
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenine and 2'-deoxyadenosine | | 分子名称: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, ACETATE ION, ADENINE, ... | | 著者 | Manszewski, T, Jaskolski, M. | | 登録日 | 2016-10-24 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
4KVO
 
 | |
4PA4
 
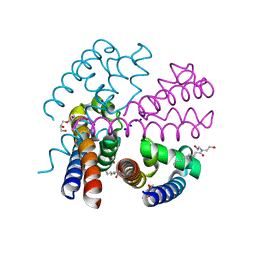 | | Structure of NavMS in complex with channel blocking compound | | 分子名称: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | 著者 | Naylor, C.E, Bagneris, C, Wallace, B.A. | | 登録日 | 2014-04-07 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5CH7
 
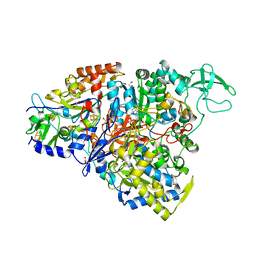 | | Crystal structure of the perchlorate reductase PcrAB - Phe164 gate switch intermediate - from Azospira suillum PS | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | 著者 | Tsai, C.-L, Tainer, J.A. | | 登録日 | 2015-07-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
2HEA
 
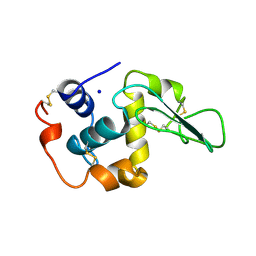 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | 登録日 | 1997-09-16 | | 公開日 | 1998-01-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2HEF
 
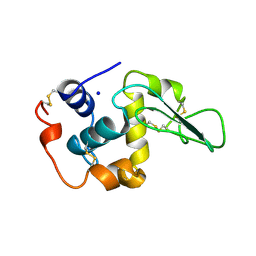 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | 登録日 | 1997-09-16 | | 公開日 | 1998-01-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2HEB
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | 登録日 | 1997-09-16 | | 公開日 | 1998-01-28 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
4NLW
 
 | |
4NLV
 
 | |
5K0J
 
 | | Crystal Structure of COMT in complex with 5-[5-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-3-yl]-2,4-dimethyl-1,3-thiazole | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-{3-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, ... | | 著者 | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | 登録日 | 2016-05-17 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
2WM2
 
 | |
4NLY
 
 | |
5MIY
 
 | | Crystal structure of the E3 ubiquitin ligase RavN from Legionella pneumophila | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin ligase RavN, SODIUM ION, ... | | 著者 | Lucas, M, Abascal-Palacios, G, Rojas, A.L, Hierro, A. | | 登録日 | 2016-11-29 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.717 Å) | | 主引用文献 | RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases.
PLoS Pathog., 14, 2018
|
|
