1OD4
 
 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | 分子名称: | ACETYL-COENZYME A CARBOXYLASE, ADENINE | | 著者 | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | 登録日 | 2003-02-12 | | 公開日 | 2003-04-03 | | 最終更新日 | 2018-06-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1OEN
 
 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | 分子名称: | ACETATE ION, PHOSPHOENOLPYRUVATE CARBOXYKINASE | | 著者 | Matte, A, Goldie, H, Sweet, R.M, Delbaere, L.T.J. | | 登録日 | 1995-09-08 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold.
J.Mol.Biol., 256, 1996
|
|
1NVT
 
 | |
1NW8
 
 | | Structure of L72P mutant beta class N6-adenine DNA methyltransferase RsrI | | 分子名称: | CHLORIDE ION, MODIFICATION METHYLASE RSRI | | 著者 | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | 登録日 | 2003-02-05 | | 公開日 | 2003-07-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NWT
 
 | | Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitopentaose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fusetti, F, Pijning, T, Kalk, K.H, Bos, E, Dijkstra, B.W. | | 登録日 | 2003-02-06 | | 公開日 | 2003-08-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39
J.Biol.Chem., 278, 2003
|
|
1ODC
 
 | | STRUCTURE OF ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH N-4'-QUINOLYL-N'-9"-(1",2",3",4"-TETRAHYDROACRIDINYL)-1,8- DIAMINOOCTANE AT 2.2A RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N-QUINOLIN-4-YL-N'-(1,2,3,4-TETRAHYDROACRIDIN-9-YL)OCTANE-1,8-DIAMINE | | 著者 | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.-F, Pang, Y.-P, Silman, I, Sussman, J.L. | | 登録日 | 2003-02-15 | | 公開日 | 2005-03-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Complexes of Alkylene-Linked Tacrine Dimers with Torpedo Californica Acetylcholinesterase: Binding of Bis(5)-Tacrine Produces a Dramatic Rearrangement in the Active-Site Gorge.
J.Med.Chem., 49, 2006
|
|
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | 分子名称: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | 著者 | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | 登録日 | 2003-02-13 | | 公開日 | 2003-03-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NSI
 
 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | 著者 | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | 登録日 | 1999-01-10 | | 公開日 | 2000-01-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
1NLK
 
 | |
1NLT
 
 | | The crystal structure of Hsp40 Ydj1 | | 分子名称: | Mitochondrial protein import protein MAS5, Seven residue peptide, ZINC ION | | 著者 | Li, J, Sha, B. | | 登録日 | 2003-01-07 | | 公開日 | 2004-01-13 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate.
Structure, 11, 2003
|
|
1NM0
 
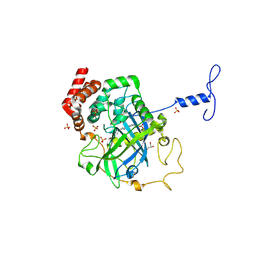 | | Proteus mirabilis catalase in complex with formiate | | 分子名称: | Catalase, FORMIC ACID, GLYCEROL, ... | | 著者 | Andreoletti, P, Pernoud, A, Gouet, P, Jouve, H.M. | | 登録日 | 2003-01-08 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural studies of Proteus mirabilis catalase in its ground state, oxidized state and in complex with formic acid.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NMT
 
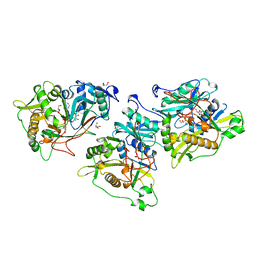 | |
1NUN
 
 | | Crystal Structure Analysis of the FGF10-FGFR2b Complex | | 分子名称: | Fibroblast growth factor-10, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | 著者 | Yeh, B.K, Igarashi, M, Eliseenkova, A.V, Plotnikov, A.N, Sher, I, Ron, D, Aaronson, S.A, Mohammadi, M. | | 登録日 | 2003-01-31 | | 公開日 | 2003-02-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis by which alternative splicing confers
specificity in fibroblast growth factor receptors.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NOY
 
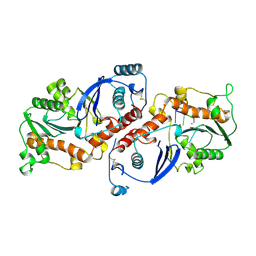 | | DNA POLYMERASE (E.C.2.7.7.7)/DNA COMPLEX | | 分子名称: | DNA (5'-D(*TP*TP*T)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE (E.C.2.7.7.7)), ... | | 著者 | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | 登録日 | 1996-02-16 | | 公開日 | 1996-10-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
1NW6
 
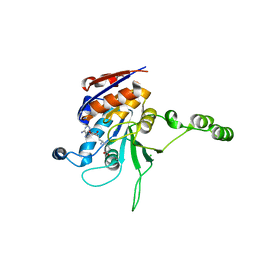 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to sinefungin | | 分子名称: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, SINEFUNGIN | | 著者 | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | 登録日 | 2003-02-05 | | 公開日 | 2003-07-29 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NVF
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+, ADP and carbaphosphonate | | 分子名称: | 3-DEHYDROQUINATE SYNTHASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | 登録日 | 2003-02-03 | | 公開日 | 2003-03-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1O7O
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-11 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | 分子名称: | Hypothetical protein ybgI, MAGNESIUM ION | | 著者 | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-10 | | 公開日 | 2004-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1O80
 
 | |
1OA7
 
 | |
1NR0
 
 | | Two Seven-Bladed Beta-Propeller Domains Revealed By The Structure Of A C. elegans Homologue Of Yeast Actin Interacting Protein 1 (AIP1). | | 分子名称: | Actin interacting protein 1, MANGANESE (II) ION | | 著者 | Vorobiev, S.M, Mohri, K, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-01-23 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Identification of Functional Residues on Caenorhabditis elegans Actin-interacting Protein 1 (UNC-78) for Disassembly of Actin Depolymerizing Factor/Cofilin-bound Actin Filaments
J.Biol.Chem., 279, 2004
|
|
1OBB
 
 | | alpha-glucosidase A, AglA, from Thermotoga maritima in complex with maltose and NAD+ | | 分子名称: | ALPHA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Lodge, J.A, Maier, T, Liebl, W, Hoffmann, V, Strater, N. | | 登録日 | 2003-01-29 | | 公開日 | 2003-05-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Thermotoga Maritima Alpha-Glucosidase Agla Defines a New Clan of Nad+-Dependent Glycosidases
J.Biol.Chem., 278, 2003
|
|
1NSN
 
 | |
1NY6
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the active state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, transcriptional regulator (NtrC family) | | 著者 | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | 登録日 | 2003-02-11 | | 公開日 | 2003-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
1O12
 
 | |
